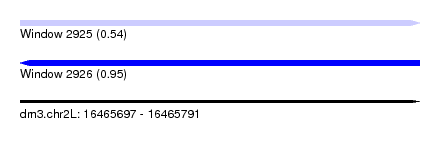
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,465,697 – 16,465,791 |
| Length | 94 |
| Max. P | 0.946041 |

| Location | 16,465,697 – 16,465,791 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.93 |
| Shannon entropy | 0.45458 |
| G+C content | 0.31704 |
| Mean single sequence MFE | -13.78 |
| Consensus MFE | -8.77 |
| Energy contribution | -8.68 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.542130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16465697 94 + 23011544 CGUACAUACGUGAUCGUUUAACUGUAAAUUAAAUAUAAUUUACCAAUAAUCAUAAUCACAAUAUGCAGUUUCUGUAUCUGAAU-CCGAUUCCGCG------------------------- ........((..((((.......((((((((....))))))))............(((....((((((...)))))).)))..-.))))..))..------------------------- ( -14.00, z-score = -1.06, R) >droEre2.scaffold_4845 19532128 94 + 22589142 CGUACAUACGUGAUCGUUUAACUGUAAAUUAAAUAUAAUUUACCAAUAAUCAUAAUCACAAUAUGCAGUUUCUGUUUCUGAAU-CCGAUUCCGCG------------------------- ........((..((((.......((((((((....))))))))............(((......((((...))))...)))..-.))))..))..------------------------- ( -12.00, z-score = -0.37, R) >droYak2.chr2R 2923969 79 + 21139217 CGUACAUACGUGAUCGUUUAACUGUAAAUUAAAUAUAAUUUACCAAUAAUCAUAAUCACAAUAUGCAGUUUCUGUUUCU----------------------------------------- .........(((((..((((...((((((((....))))))))...)))..)..))))).....((((...))))....----------------------------------------- ( -9.70, z-score = -0.85, R) >droSec1.super_7 111868 94 + 3727775 CGUACAUACGUGAUCGUUUAACUGUAAAUUAAAUAUAAUUUACCAAUAAUCAUAAUCACAAUAUGCAGUUUCUGUAUCUGAAU-CCGAUUCCGCG------------------------- ........((..((((.......((((((((....))))))))............(((....((((((...)))))).)))..-.))))..))..------------------------- ( -14.00, z-score = -1.06, R) >droSim1.chr3R 21668029 94 - 27517382 CGUACAUACGUGAUCGUUUAACUGUAAAUUAAAUAUAAUUUACCAAUAAUCAUAAUCACAAUAUGCAGUUUCUGUAUCUGAAU-CCGAUUCCGCG------------------------- ........((..((((.......((((((((....))))))))............(((....((((((...)))))).)))..-.))))..))..------------------------- ( -14.00, z-score = -1.06, R) >droAna3.scaffold_12916 93967 102 - 16180835 CGCACAUACGUGAUCGUUUAACUGUAAAUUAAAUAUAAUUUACCAAUAAUCAUAAUCACAAUAUGCAGUUUCUGUAUCUGUUU--CGCUUUUGCUCCUCUUGUA---------------- .(((.....(((((..((((...((((((((....))))))))...)))..)..)))))...((((((...))))))......--......)))..........---------------- ( -14.40, z-score = -1.24, R) >dp4.chr4_group3 8188070 120 + 11692001 CGUACAUACGUGAUCGUUUAACUGUAAAUUAAAUAUAAUUUACCAAUAAUCAUAAUCACAAUAUGCAGUUUCUGUUUCUGUGUGCUGCCUCUGCCUCUCUUCUCUCUCCUUUCUGCCGCG .(((((((.(((((..((((...((((((((....))))))))...)))..)..))))).....((((...))))...)))))))................................... ( -16.90, z-score = -2.32, R) >droPer1.super_1 9637949 120 + 10282868 CGUACAUACGUGAUCGUUUAACUGUAAAUUAAAUAUAAUUUACCAAUAAUCAUAAUCACAAUAUGCAGUUUCUGUUUCUGUGUGCUGCCUCUGCCUCUCUUCUCUCUCCUUUCUGCCGCG .(((((((.(((((..((((...((((((((....))))))))...)))..)..))))).....((((...))))...)))))))................................... ( -16.90, z-score = -2.32, R) >droWil1.scaffold_180772 4681158 104 + 8906247 CGCACAUACGUGAUCGUUUAACUGUAAAUUAAAUAUAAUUUACCAAUAAUCAUAAUCACAAUAUAUCGUAUAUGCAUAUAGUUUCUGUCUCUGUUCUUUUGUUA---------------- .(((.(((((((((..((((...((((((((....))))))))...)))..)..)))))........)))).))).............................---------------- ( -12.60, z-score = -0.25, R) >droVir3.scaffold_12963 7192472 97 - 20206255 CGCGCAUACGUGAUCGUUUAACUGUAAAUUAAAUAUAAUUUACCAAUAAGUAUAAUUACAAUACGUAGUUU-------UGUUUUUUUGUUGUGUUCCUUUGGUU---------------- ((((....)))).......((((((((((((....))))))))(((((((...((((((.....)))))).-------......))))))).........))))---------------- ( -13.30, z-score = 0.57, R) >consensus CGUACAUACGUGAUCGUUUAACUGUAAAUUAAAUAUAAUUUACCAAUAAUCAUAAUCACAAUAUGCAGUUUCUGUAUCUGAAU_CCGAUUCUGCG_________________________ .........(((((..((((...((((((((....))))))))...)))..)..)))))............................................................. ( -8.77 = -8.68 + -0.09)

| Location | 16,465,697 – 16,465,791 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.93 |
| Shannon entropy | 0.45458 |
| G+C content | 0.31704 |
| Mean single sequence MFE | -18.00 |
| Consensus MFE | -15.95 |
| Energy contribution | -15.85 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
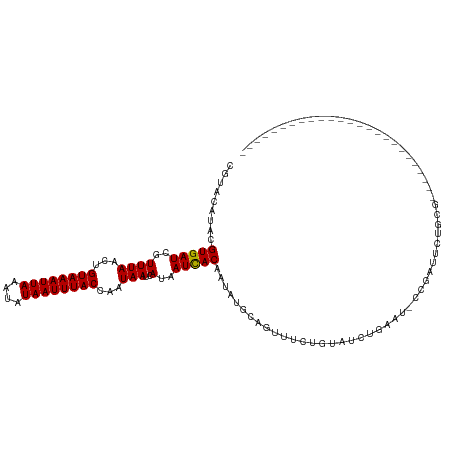
>dm3.chr2L 16465697 94 - 23011544 -------------------------CGCGGAAUCGG-AUUCAGAUACAGAAACUGCAUAUUGUGAUUAUGAUUAUUGGUAAAUUAUAUUUAAUUUACAGUUAAACGAUCACGUAUGUACG -------------------------.((((..((..-...........))..))))((((((((((..(....((((.(((((((....)))))))))))...)..)))))).))))... ( -17.72, z-score = -0.73, R) >droEre2.scaffold_4845 19532128 94 - 22589142 -------------------------CGCGGAAUCGG-AUUCAGAAACAGAAACUGCAUAUUGUGAUUAUGAUUAUUGGUAAAUUAUAUUUAAUUUACAGUUAAACGAUCACGUAUGUACG -------------------------.((((..((..-...........))..))))((((((((((..(....((((.(((((((....)))))))))))...)..)))))).))))... ( -17.72, z-score = -0.89, R) >droYak2.chr2R 2923969 79 - 21139217 -----------------------------------------AGAAACAGAAACUGCAUAUUGUGAUUAUGAUUAUUGGUAAAUUAUAUUUAAUUUACAGUUAAACGAUCACGUAUGUACG -----------------------------------------............(((((((.(((((..(....((((.(((((((....)))))))))))...)..)))))))))))).. ( -17.30, z-score = -2.57, R) >droSec1.super_7 111868 94 - 3727775 -------------------------CGCGGAAUCGG-AUUCAGAUACAGAAACUGCAUAUUGUGAUUAUGAUUAUUGGUAAAUUAUAUUUAAUUUACAGUUAAACGAUCACGUAUGUACG -------------------------.((((..((..-...........))..))))((((((((((..(....((((.(((((((....)))))))))))...)..)))))).))))... ( -17.72, z-score = -0.73, R) >droSim1.chr3R 21668029 94 + 27517382 -------------------------CGCGGAAUCGG-AUUCAGAUACAGAAACUGCAUAUUGUGAUUAUGAUUAUUGGUAAAUUAUAUUUAAUUUACAGUUAAACGAUCACGUAUGUACG -------------------------.((((..((..-...........))..))))((((((((((..(....((((.(((((((....)))))))))))...)..)))))).))))... ( -17.72, z-score = -0.73, R) >droAna3.scaffold_12916 93967 102 + 16180835 ----------------UACAAGAGGAGCAAAAGCG--AAACAGAUACAGAAACUGCAUAUUGUGAUUAUGAUUAUUGGUAAAUUAUAUUUAAUUUACAGUUAAACGAUCACGUAUGUGCG ----------------..........((....)).--.................((((((((((((..(....((((.(((((((....)))))))))))...)..)))))).)))))). ( -21.00, z-score = -2.25, R) >dp4.chr4_group3 8188070 120 - 11692001 CGCGGCAGAAAGGAGAGAGAAGAGAGGCAGAGGCAGCACACAGAAACAGAAACUGCAUAUUGUGAUUAUGAUUAUUGGUAAAUUAUAUUUAAUUUACAGUUAAACGAUCACGUAUGUACG .((.((..........................)).))................(((((((.(((((..(....((((.(((((((....)))))))))))...)..)))))))))))).. ( -19.57, z-score = -0.78, R) >droPer1.super_1 9637949 120 - 10282868 CGCGGCAGAAAGGAGAGAGAAGAGAGGCAGAGGCAGCACACAGAAACAGAAACUGCAUAUUGUGAUUAUGAUUAUUGGUAAAUUAUAUUUAAUUUACAGUUAAACGAUCACGUAUGUACG .((.((..........................)).))................(((((((.(((((..(....((((.(((((((....)))))))))))...)..)))))))))))).. ( -19.57, z-score = -0.78, R) >droWil1.scaffold_180772 4681158 104 - 8906247 ----------------UAACAAAAGAACAGAGACAGAAACUAUAUGCAUAUACGAUAUAUUGUGAUUAUGAUUAUUGGUAAAUUAUAUUUAAUUUACAGUUAAACGAUCACGUAUGUGCG ----------------.............................((((((((((....))(((((..(....((((.(((((((....)))))))))))...)..))))))))))))). ( -20.00, z-score = -2.07, R) >droVir3.scaffold_12963 7192472 97 + 20206255 ----------------AACCAAAGGAACACAACAAAAAAACA-------AAACUACGUAUUGUAAUUAUACUUAUUGGUAAAUUAUAUUUAAUUUACAGUUAAACGAUCACGUAUGCGCG ----------------..........................-------....((((((((((.......(.....)((((((((....))))))))......))))).)))))...... ( -11.70, z-score = -0.24, R) >consensus _________________________CGCAGAAUCGG_AUACAGAUACAGAAACUGCAUAUUGUGAUUAUGAUUAUUGGUAAAUUAUAUUUAAUUUACAGUUAAACGAUCACGUAUGUACG .....................................................(((((((.((((((....(((...((((((((....))))))))...)))..))))))))))))).. (-15.95 = -15.85 + -0.10)
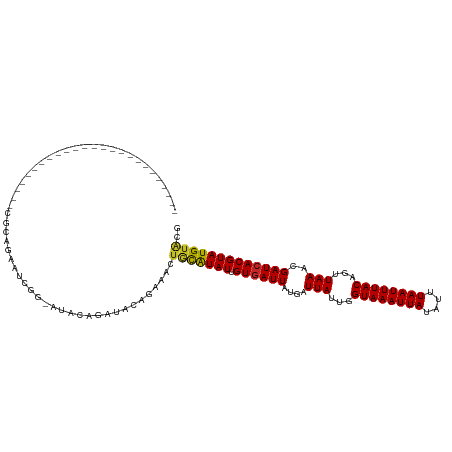
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:17 2011