| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,464,577 – 16,464,710 |
| Length | 133 |
| Max. P | 0.911511 |

| Location | 16,464,577 – 16,464,674 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.21 |
| Shannon entropy | 0.44929 |
| G+C content | 0.54863 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -13.80 |
| Energy contribution | -15.80 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
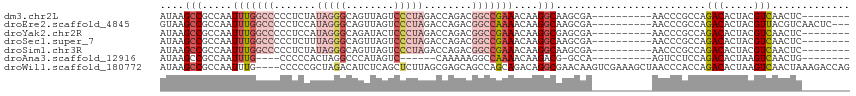
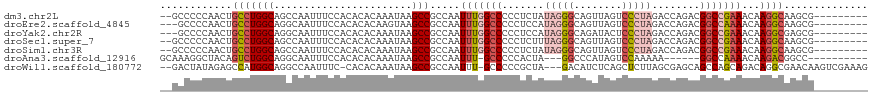
>dm3.chr2L 16464577 97 - 23011544 AUAAGCCGCCAAUUUGGCCCCCUCUAUAGGGCAGUUAGUCCCUAGACCAGACGGCCGAAACAAGGCAAGCGA----------AACCCGCCAGACACUACGUCAACUC-------- ....(((.....(((((((...(((.((((((.....).)))))....))).)))))))....)))..(((.----------....)))..(((.....))).....-------- ( -27.80, z-score = -2.19, R) >droEre2.scaffold_4845 19531100 102 - 22589142 GUAAGCCGCCAAUUUGGCCCCCUCCAUAGGGCAGUUAGUCCCUAGACCAGACGGCCAAAACAAGGCAAGCGA----------AACCCGCCAGACACUACGUUACGUCAACUC--- (((((((.....(((((((((((....))))..(((.(((....)))..))))))))))....)))..(((.----------....)))...........))))........--- ( -27.50, z-score = -1.91, R) >droYak2.chr2R 2922806 97 - 21139217 AUAAGCCGCCAAUUUGGCCCCCUCCAUAGGGCAGAUACUCCCUAGACCAGACGGCCGAAACAAGGCGAGCGA----------AACCCGCCAGACACUACGUCAACUC-------- ....((((((..(((((((..((...(((((.((...)))))))....))..)))))))....)))).))..----------.........(((.....))).....-------- ( -28.40, z-score = -2.76, R) >droSec1.super_7 110752 97 - 3727775 AUAAGCCGCCAAUUUGGCCCCCUCUUUAGGGCAGUUAGUCCCUAGACCAGACGGCCGAAACAAGGCAAGCGA----------AACCCGCCAGACACUACGUCAACUC-------- ....(((.....(((((((..((.((((((((.....).)))))))..))..)))))))....)))..(((.----------....)))..(((.....))).....-------- ( -28.40, z-score = -2.29, R) >droSim1.chr3R 21666913 97 + 27517382 AUAAGCCGCCAAUUUGGCCCCCUCUAUAGGGCAGUUAGUCCCUAGACCAGACGGCCGAAACAAGGCAAGCGA----------AACCCGCCAGACACUACGUCAACUC-------- ....(((.....(((((((...(((.((((((.....).)))))....))).)))))))....)))..(((.----------....)))..(((.....))).....-------- ( -27.80, z-score = -2.19, R) >droAna3.scaffold_12916 92971 86 + 16180835 AUAAGCCGCCAAUUUG----CCCCCACUAGGCCCAUAGUC------CAAAAAGGCCAAAACAAGACG-GCCA----------AGUCCUCCAGACACUAAGUCAACUG-------- ....(((((......)----)........((((.......------......))))..........)-))..----------.........(((.....))).....-------- ( -13.52, z-score = -0.37, R) >droWil1.scaffold_180772 4678164 111 - 8906247 AUAAGCCGCCAAUUUG----CCCCCGCUAGACAUCUCAGCUCUUAGCGAGCAGCCAGCAGACAGGCGAACAAGUCGAAAGCUAACCCACCAGACACUAAGUCAACUAAAGACCAG ......((((..((((----(....(((.((....)).((((.....)))))))..)))))..))))....((.(....))).........(((.....)))............. ( -23.20, z-score = -1.75, R) >consensus AUAAGCCGCCAAUUUGGCCCCCUCCAUAGGGCAGUUAGUCCCUAGACCAGACGGCCGAAACAAGGCAAGCGA__________AACCCGCCAGACACUACGUCAACUC________ ....(((.....(((((((.......(((((........)))))........)))))))....))).........................(((.....)))............. (-13.80 = -15.80 + 2.00)
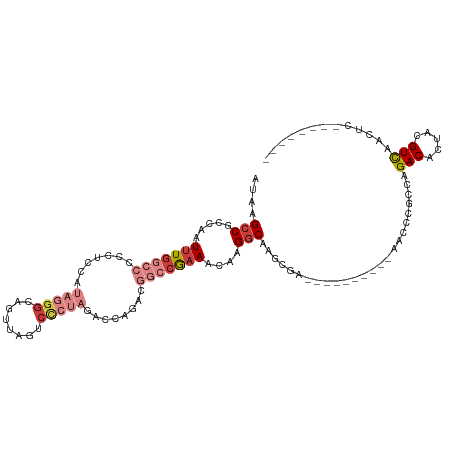
| Location | 16,464,603 – 16,464,710 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.08 |
| Shannon entropy | 0.43719 |
| G+C content | 0.56037 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -16.21 |
| Energy contribution | -17.50 |
| Covariance contribution | 1.29 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.786897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
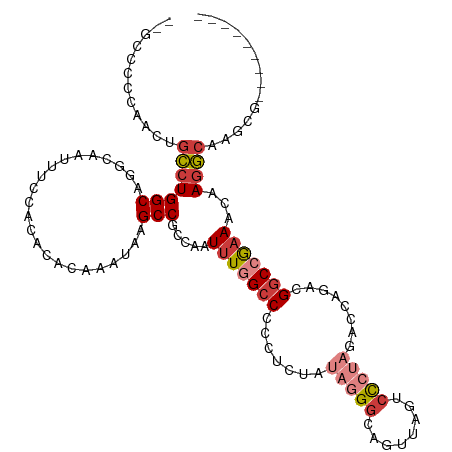
>dm3.chr2L 16464603 107 - 23011544 --GCCCCCAACUGCCUGGCAGCCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCUAUAGGGCAGUUAGUCCCUAGACCAGACGGCCGAAACAAGGCAAGCG--------- --.........((((((((.((..((((........))))..)).)))..(((((((...(((.((((((.....).)))))....))).)))))))...)))))....--------- ( -31.30, z-score = -1.95, R) >droEre2.scaffold_4845 19531131 106 - 22589142 ---GCCCCAACUGCCUGGCAGGCAAUUUCCACACACAAGUAAGCCGCCAAUUUGGCCCCCUCCAUAGGGCAGUUAGUCCCUAGACCAGACGGCCAAAACAAGGCAAGCG--------- ---........((((((((.(((...................))))))..(((((((((((....))))..(((.(((....)))..))))))))))...)))))....--------- ( -32.51, z-score = -1.64, R) >droYak2.chr2R 2922832 106 - 21139217 ---GCCCCAACUGCCUGGCAGGCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCCAUAGGGCAGAUACUCCCUAGACCAGACGGCCGAAACAAGGCGAGCG--------- ---((((((......)))..)))...................((((((..(((((((..((...(((((.((...)))))))....))..)))))))....)))).)).--------- ( -32.10, z-score = -1.94, R) >droSec1.super_7 110778 107 - 3727775 --GCCCCCAACUGCCUGGCAGCCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCUUUAGGGCAGUUAGUCCCUAGACCAGACGGCCGAAACAAGGCAAGCG--------- --.........((((((((.((..((((........))))..)).)))..(((((((..((.((((((((.....).)))))))..))..)))))))...)))))....--------- ( -31.90, z-score = -2.03, R) >droSim1.chr3R 21666939 107 + 27517382 --GCCCCCAACUGCCUGGCAGCCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCUAUAGGGCAGUUAGUCCCUAGACCAGACGGCCGAAACAAGGCAAGCG--------- --.........((((((((.((..((((........))))..)).)))..(((((((...(((.((((((.....).)))))....))).)))))))...)))))....--------- ( -31.30, z-score = -1.95, R) >droAna3.scaffold_12916 92997 98 + 16180835 GCAAAGGCUACAGUCUGGCAGGCAAUUUCCACACACAAAUAAGCCGCCAAUUU-GCCCCCACUA---GGCCCAUAGUCCAAAAA------GGCCAAAACAAGACGGCC---------- .....((((...(((((((.(((...................)))))).....-..........---((((.............------))))......))))))))---------- ( -22.03, z-score = -0.24, R) >droWil1.scaffold_180772 4678199 111 - 8906247 --GACUAUAGAGCCAUGGCAGGCCAAUUUC-CACACAAAUAAGCCGCCAAUUU-GCCCCCGCUA---GACAUCUCAGCUCUUAGCGAGCAGCCAGCAGACAGGCGAACAAGUCGAAAG --((((.....(((.((((.(((..((((.-.....))))..))))))).(((-((....(((.---((....)).((((.....)))))))..)))))..))).....))))..... ( -31.30, z-score = -2.81, R) >consensus __GCCCCCAACUGCCUGGCAGGCAAUUUCCACACACAAAUAAGCCGCCAAUUUGGCCCCCUCUAUAGGGCAGUUAGUCCCUAGACCAGACGGCCGAAACAAGGCAAGCG_________ ............(((((((.......................))).....(((((((.......(((((........)))))........)))))))...)))).............. (-16.21 = -17.50 + 1.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:15 2011