
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,460,201 – 16,460,300 |
| Length | 99 |
| Max. P | 0.661107 |

| Location | 16,460,201 – 16,460,300 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.45 |
| Shannon entropy | 0.41939 |
| G+C content | 0.28893 |
| Mean single sequence MFE | -12.73 |
| Consensus MFE | -7.89 |
| Energy contribution | -8.22 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
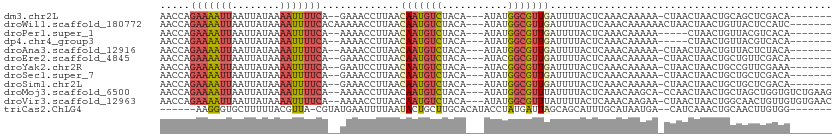
>dm3.chr2L 16460201 99 + 23011544 AACCAGAAAAUUAAUUAUAAAAUUUUCA--GAAACCUUAACAAUGUCUACA---AUAUGGCGUUGAUUUUACUCAAACAAAAA-CUAACUAACUGCAGCUCGACA------- .....(((((((........))))))).--..........(((((((....---....)))))))..................-.....................------- ( -11.70, z-score = -1.39, R) >droWil1.scaffold_180772 4669565 102 + 8906247 AACCAGAAAAUUAAUUAUAAAAUUUUCACAAAAACCUUAACAAUGUCUACA---AUAUGGCGUUGAUUUUACUCAAACAAAAAACUAACUAACUGUUACUCCAUC------- .....(((((((........))))))).............(((((((....---....)))))))....................((((.....)))).......------- ( -12.60, z-score = -2.67, R) >droPer1.super_1 9632369 95 + 10282868 AACCAGAAAAUUAAUUAUAAAAUUUUCA--AAAACCUUAACAAUGUCUACA---AUAUGGCGUUGAUUUUACUCAAACAAAAA-----CUAACUGUUACGUCACA------- .....(((((((........))))))).--..........(((((((....---....)))))))..................-----.................------- ( -11.70, z-score = -1.56, R) >dp4.chr4_group3 8182443 95 + 11692001 AACCAGAAAAUUAAUUAUAAAAUUUUCA--AAAACCUUAACAAUGUCUACA---AUAUGGCGUUGAUUUUACUCAAACAAAAA-----CUAACUGUUACGUCACA------- .....(((((((........))))))).--..........(((((((....---....)))))))..................-----.................------- ( -11.70, z-score = -1.56, R) >droAna3.scaffold_12916 88364 99 - 16180835 AACCAGAAAAUUAAUUAUAAAAUUUUCA--AAAACCUUAACAAUGUCUACA---AUAUGGCGUUGAUUUUACUCAAACAAAAA-CUAACUAACUGUUACUCUACA------- .....(((((((........))))))).--..........(((((((....---....)))))))..................-.((((.....)))).......------- ( -12.60, z-score = -2.82, R) >droEre2.scaffold_4845 19526847 99 + 22589142 AACCAGAAAAUUAAUUAUAAAAUUUUCA--GAAACCUUAACAAUGUCUACA---AUACGGCGUUGAUUUUACUCAAACAAAAA-CUAACUAACUGCUGUUCGACA------- .....(((((((........))))))).--..........(((((((....---....))))))).......((.((((....-............)))).))..------- ( -11.19, z-score = -1.04, R) >droYak2.chr2R 2917533 99 + 21139217 AACCAGAAAAUUAAUUAUAAAAUUUUCA--GAAUCCUUAACAAUGUCUACA---AUACGGCGUUGAUUUUACUCAAACAAAAA-CUAACUAACUGCCGUUCGAAA------- .....(((((((........))))))).--..........(((((((....---....)))))))..................-.....................------- ( -11.10, z-score = -1.05, R) >droSec1.super_7 106337 99 + 3727775 AACCAGAAAAUUAAUUAUAAAAUUUUCA--GAAACCUUAACAAUGUCUACA---AUAUGGCGUUGAUUUUACUCAAACAAAAA-CUAACUAACUGCUGCUCGACA------- .....(((((((........))))))).--..........(((((((....---....)))))))..................-.....................------- ( -11.70, z-score = -1.52, R) >droSim1.chr2L 16172419 99 + 22036055 AACCAGAAAAUUAAUUAUAAAAUUUUCA--GAAACCUUAACAAUGUCUACA---AUAUGGCGUUGAUUUUACUCAAACAAAAA-CUAACUAACUGCUGCUCGACA------- .....(((((((........))))))).--..........(((((((....---....)))))))..................-.....................------- ( -11.70, z-score = -1.52, R) >droMoj3.scaffold_6500 7063449 106 + 32352404 AACCAGAAAAUUAAUUAUAAAAUUUUCA--AAAACCUUAACAAUGUCUACA---AUAUGGCGUUUAUUUUACUCAAACAAGCA-CCAACUAACUGCUAGCUGGUGUCUGAAG .....(((((((........))))))).--...........((((((....---....)))))).............((..((-(((.(((.....))).)))))..))... ( -19.70, z-score = -2.71, R) >droVir3.scaffold_12963 7187132 106 - 20206255 AACCAGAAAAUUAAUUAUAAAAUUUUCA--AAAACCUUAACAAUGUCUACA---AUAUGGCGUUUAUUUUACUCAAACAAGAA-CUAACUAACUGGCAACUGUUGUGUGAAC .....(((((((........))))))).--...........((((((....---....))))))...(((((.(((...((..-(((......)))...)).))).))))). ( -12.90, z-score = -0.17, R) >triCas2.ChLG4 2430523 96 + 13894384 ------AAGGGUGCUUUUUACGUUA-CGUAUGAAUUUUAAUACUGCUUGCACAUACCUAUGAUUAGCAGCAUUUGCAUAAUGA--CAUCAAACUGCAACUUGUGG------- ------.(((.(((..(((..((((-..((((......(((.(((((..((........))...))))).)))..)))).)))--)...)))..))).)))....------- ( -14.20, z-score = 1.49, R) >consensus AACCAGAAAAUUAAUUAUAAAAUUUUCA__AAAACCUUAACAAUGUCUACA___AUAUGGCGUUGAUUUUACUCAAACAAAAA_CUAACUAACUGCUACUCGACA_______ .....(((((((........))))))).............(((((((...........)))))))............................................... ( -7.89 = -8.22 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:14 2011