
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,448,966 – 16,449,062 |
| Length | 96 |
| Max. P | 0.701488 |

| Location | 16,448,966 – 16,449,062 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 64.80 |
| Shannon entropy | 0.70999 |
| G+C content | 0.56367 |
| Mean single sequence MFE | -26.28 |
| Consensus MFE | -10.80 |
| Energy contribution | -11.69 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.59 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
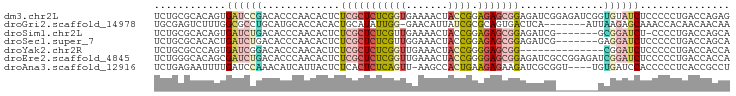
>dm3.chr2L 16448966 96 - 23011544 UCUGCGCACAGUGAUCCGACACCCAACACUCUCGCUCUCGGUGAAAACUACCGGAGAGCGGAGAUCGGAGAUCGGUGUAUCUCCCCCUGACCAGAG ((((.(..(((.(...............(((.(((((((((((.....)))).))))))))))...((((((......)))))).)))).))))). ( -32.20, z-score = -1.26, R) >droGri2.scaffold_14978 654875 88 - 1124632 UGCGAGUCUUUGGCGCCUGCAUGCACCACACUGCAUAUUGG-GAACAUUAUCGCGCAGUGACUCA-------AUUAAGAGCAAACCACAACAACAA ...(((((..(((.((......)).))).((((((((.((.-...)).)))...)))))))))).-------........................ ( -20.70, z-score = -0.50, R) >droSim1.chr2L 16161146 88 - 22036055 UCUGCGCACAGUGAUCUGACACCCAACACUCUCGCUCUCGUUGAAAACUACCGGAGAGCGGAGAUCG-------GCGGAUCU-CCCCUGACCAGCA ...((...(((.((((((....((....(((.(((((((..............))))))))))...)-------))))))).-...)))....)). ( -25.04, z-score = -0.55, R) >droSec1.super_7 95052 89 - 3727775 UCUGCGCACACUGAUCUGACACCCAACACUCUCGCUCUCGUUGGAAACUACCGGAGAGCGGAGAUCG-------GAGGAUCUCCCCCUGACCAGCA ..........(((.((.................((((((((.(....).))..))))))(((((((.-------...)))))))....)).))).. ( -26.30, z-score = -0.93, R) >droYak2.chr2R 2906054 82 - 21139217 UCUGCGCCCAGUGAUCGGACACCCAACACUCUCGCUCUCGGUUGAAACUACCGGGGAGCGG--------------CGGAUCUCCCCCUGACCACCA (((((....((((...((....))..))))..((((((((((.......))).))))))))--------------))))................. ( -24.00, z-score = -0.05, R) >droEre2.scaffold_4845 19515684 96 - 22589142 UCUGGGCACAGCGAUCUGACACCCAACACUCUCGCUCUCGGUUGAAACUACCGGGGAGCGGAGAUCGCCGGAGAUCGGAUCUCCCCCUGACCACCA ((.(((....((((((((.....).......(((((((((((.......))).))))))))))))))).((((((...))))))))).))...... ( -34.10, z-score = -1.29, R) >droAna3.scaffold_12916 77346 91 + 16180835 UCUGAGAAUUUUGAUCCAAACAUCAUUACUCUCACUCUCAGUU-AAGCCACUGAAGAGAAGAUCGCGGU----UGUGAUCCACCCCCUCACCGCCU ..(((((.(..((((......))))..).)))))((((((((.-.....)))).))))........(((----.((((.........)))).))). ( -21.60, z-score = -2.34, R) >consensus UCUGCGCACAGUGAUCCGACACCCAACACUCUCGCUCUCGGUGAAAACUACCGGAGAGCGGAGAUCG_______GCGGAUCUCCCCCUGACCACCA ............((((((.............(((((((((((.......)))).)))))))..............))))))............... (-10.80 = -11.69 + 0.88)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:13 2011