
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,434,479 – 16,434,581 |
| Length | 102 |
| Max. P | 0.896890 |

| Location | 16,434,479 – 16,434,581 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.87 |
| Shannon entropy | 0.28279 |
| G+C content | 0.36042 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -21.51 |
| Energy contribution | -21.76 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896890 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
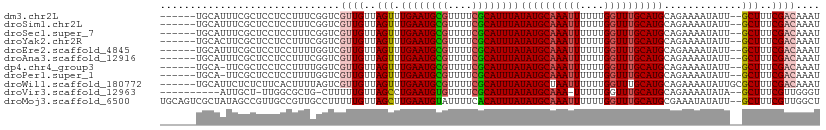
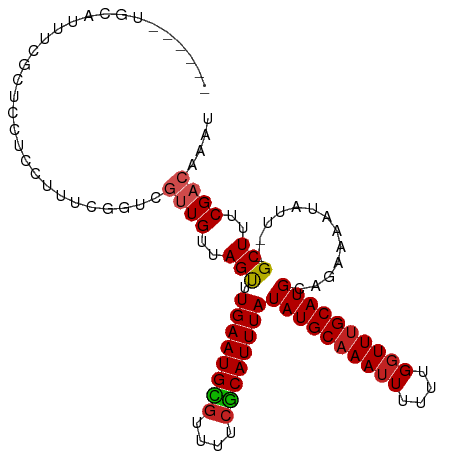
>dm3.chr2L 16434479 102 - 23011544 ------UGCAUUUCGCUCCUCCUUUCGGUCGUUGUUAGUUUGAAUGCGUUUUCGCAUUUAUAUGCAAAUUUUUUGGUUUGCAUGCAGAAAAUAUU--GCUUUCGACAAAU ------.((.....))..............((((..(((.((((((((....))))))))((((((((((....))))))))))...........--)))..)))).... ( -24.20, z-score = -1.61, R) >droSim1.chr2L 16146455 102 - 22036055 ------UGCAUUUCGCUCCUCCUUUCGGUCGUUGUUAGUUUGAAUGCGUUUUCGCAUUUAUAUGCAAAUUUUUUGGUUUGCAUGCAGAAAAUAUU--GCUUUCGACAAAU ------.((.....))..............((((..(((.((((((((....))))))))((((((((((....))))))))))...........--)))..)))).... ( -24.20, z-score = -1.61, R) >droSec1.super_7 80255 102 - 3727775 ------UGCAUUUCGCUCCUCCUUUCGGUCGUUGUUAGUUUGAAUGCGUUUUCGCAUUUAUAUGCAAAUUUUUUGGUUUGCAUGCAGAAAAUAUU--GCUUUCGACAAAU ------.((.....))..............((((..(((.((((((((....))))))))((((((((((....))))))))))...........--)))..)))).... ( -24.20, z-score = -1.61, R) >droYak2.chr2R 2891646 102 - 21139217 ------UGCACUUCGCUCCUCCUUUCGGUCGUUGUUAGUUUGAAUGCGUUUUCGCAUUUAUAUGCAAAUUUUUUGGUUUGCAUGCAGAAAAUAUU--GCUUUCGACAAAU ------.((.....))..............((((..(((.((((((((....))))))))((((((((((....))))))))))...........--)))..)))).... ( -24.20, z-score = -1.60, R) >droEre2.scaffold_4845 19498621 102 - 22589142 ------UGCAUUUCGCUCCUCCUUUUGGUCGUUGUUAGUUUGAAUGCGUUUUCGCAUUUAUAUGCAAAUUUUUUGGUUUGCAUGCAGAAAAUAUU--GCUUUCGACAAAU ------.((.....))....((....))..((((..(((.((((((((....))))))))((((((((((....))))))))))...........--)))..)))).... ( -24.60, z-score = -1.78, R) >droAna3.scaffold_12916 63454 102 + 16180835 ------UGCAUUUCGCUCCUCCUUUCGGUCGUUGUUAGUUUGAAUGCGUUUUCGCAUUUAUAUGCAAAUUUUUUGGUUUGCAUGCAGAAAAUAUU--GCUUUCGACAAAU ------.((.....))..............((((..(((.((((((((....))))))))((((((((((....))))))))))...........--)))..)))).... ( -24.20, z-score = -1.61, R) >dp4.chr4_group3 8158974 101 - 11692001 ------UGCA-UUCGCUCCUCCUUUUGGUCGUUGUUAGUUUGAAUGCGUUUUCGCAUUUAUAUGCAAAUUUUUUGGUUUGCAUGCAGAAAAUAUU--GCUUUCGACAAAU ------.((.-...))....((....))..((((..(((.((((((((....))))))))((((((((((....))))))))))...........--)))..)))).... ( -24.60, z-score = -1.76, R) >droPer1.super_1 9609062 101 - 10282868 ------UGCA-UUCGCUCCUCCUUUUGGUCGUUGUUAGUUUGAAUGCGUUUUCGCAUUUAUAUGCAAAUUUUUUGGUUUGCAUGCAGAAAAUAUU--GCUUUCGACAAAU ------.((.-...))....((....))..((((..(((.((((((((....))))))))((((((((((....))))))))))...........--)))..)))).... ( -24.60, z-score = -1.76, R) >droWil1.scaffold_180772 4622069 104 - 8906247 ------UGCAUUCUCUCUUCACUUUUAGUCGUUGUUAGUUUGAAUGCGUUUUCGCAUUUAUAUGCUAAUUUUUUGGUUUGCAUGCAGAAAAUAUUGCGCUUUCGACAAAU ------.....................((((..((((((.((((((((....))))))))...))))((((((((.........)))))))).....))...)))).... ( -20.90, z-score = -0.69, R) >droVir3.scaffold_12963 7153760 95 + 20206255 ----------AUUGCU-UUGGCGCUG-CUUUUUGUUAGCCUGAAUGUGUUUUCGCAUUUAUAUGCAAA-UUUUUGGUUUGCAUGCAGAAAAUAUA--GCUUUCGUUGGGU ----------.....(-(..(((..(-((((((((.....((((((((....)))))))).(((((((-(.....)))))))))))))).....)--))...)))..)). ( -23.70, z-score = -0.69, R) >droMoj3.scaffold_6500 7037431 108 - 32352404 UGCAGUCGCUAUAGCCGUUGCCGUUGCCUUUUUGUUAGCUUGAAUGUAUUUUCACAUUUAUAUGCAAAUUUUUUGGUUUGCAUGCGAAAUAUAUU--GCUUUCGUUGGCU .((((..((....))..))))....(((....((..(((.(((((((......)))))))((((((((((....))))))))))...........--)))..))..))). ( -27.10, z-score = -1.29, R) >consensus ______UGCAUUUCGCUCCUCCUUUCGGUCGUUGUUAGUUUGAAUGCGUUUUCGCAUUUAUAUGCAAAUUUUUUGGUUUGCAUGCAGAAAAUAUU__GCUUUCGACAAAU ..............................((((..(((.((((((((....))))))))((((((((((....)))))))))).............)))..)))).... (-21.51 = -21.76 + 0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:10 2011