
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,413,878 – 16,413,979 |
| Length | 101 |
| Max. P | 0.941451 |

| Location | 16,413,878 – 16,413,979 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 92.83 |
| Shannon entropy | 0.12352 |
| G+C content | 0.47729 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -26.36 |
| Energy contribution | -27.16 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.941451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
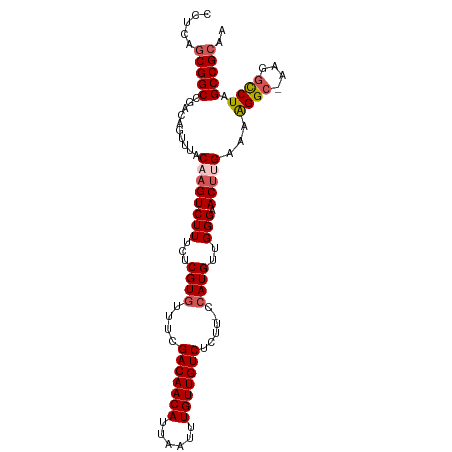
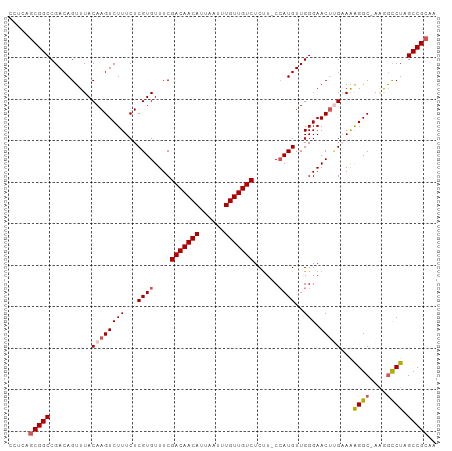
>dm3.chr2L 16413878 101 + 23011544 CCUCAGCGGCCGACAGUUUACAAGUCUUUCUCGUGUUUCGACAACAUUAAUUUGUUGUCUCUU-CCAUGUUGGGAACUUGAAAAGGC-AAGGCCUAGCCGCAA .....(((((..........((((((((...((((....(((((((......)))))))....-.))))..))).)))))...((((-...)))).))))).. ( -32.50, z-score = -2.92, R) >droEre2.scaffold_4845 19478743 103 + 22589142 CCUCAGCGGCCGACAGUUUACAAGUCUUUCUCGUGUUUCGACAACAUUAAUUUGUUGUCUCCUGCAAUGUUGGGAACGCGAAAGGGCCAAAACCUAGCCGCAU .....(((((.....((((....((((((.((((((((((((((((......))))))).....((....)))))))))))))))))..))))...))))).. ( -35.20, z-score = -3.30, R) >droYak2.chr2R 2870743 101 + 21139217 CCUCAGCGGCCGACAGUUUACAAGUCUUUCUCGUGUUUCGACAACAUUAAUUUGUUGUCUCUU-CCAUGUUGGGAACUGGAAAGGGC-AGAGCCUAGCCGCAU .....(((((...(((((..(((........((((....(((((((......)))))))....-.)))))))..)))))....((((-...)))).))))).. ( -31.80, z-score = -2.04, R) >droSec1.super_7 59604 101 + 3727775 CCUCAGCGGCCGACAGUUUACAAGUCUUUCUCGUGUUUCGACAACAUUAAUUUGUUGUCUCUU-CCAUGUUGGGAACUUGCAGAGGC-AAGGCCUAGCCGCAA .....((((((((((...(((.((.....)).)))....(((((((......)))))))....-...)))))((..(((((....))-)))..)).))))).. ( -32.70, z-score = -2.56, R) >droSim1.chr2L 16125115 101 + 22036055 CCUCAACGGCCGACAGUUUACAAGUCUUUCUCGUGUUUCGACAACAUUAAUUUGUUGUCUCUU-CCAUGUUGGGAACUUGCAGAGGC-AAGGUCUAGCCGCAA ......((((.(((..........((((...((((....(((((((......)))))))....-.))))..)))).(((((....))-))))))..))))... ( -28.80, z-score = -1.87, R) >consensus CCUCAGCGGCCGACAGUUUACAAGUCUUUCUCGUGUUUCGACAACAUUAAUUUGUUGUCUCUU_CCAUGUUGGGAACUUGAAAAGGC_AAGGCCUAGCCGCAA .....(((((..........((((((((...((((....(((((((......)))))))......))))..))).)))))...((((....)))).))))).. (-26.36 = -27.16 + 0.80)

| Location | 16,413,878 – 16,413,979 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Shannon entropy | 0.12352 |
| G+C content | 0.47729 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -24.90 |
| Energy contribution | -24.98 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
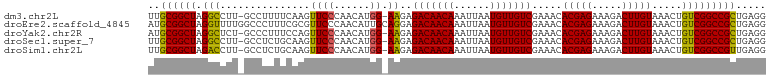
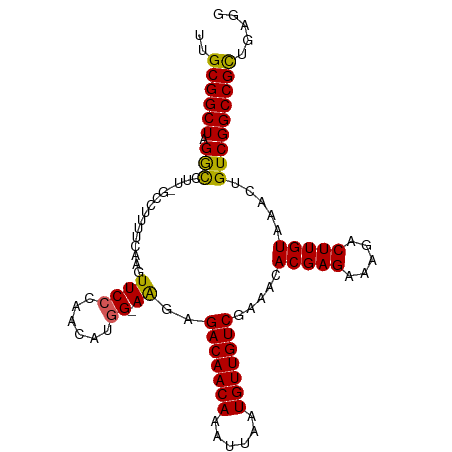
>dm3.chr2L 16413878 101 - 23011544 UUGCGGCUAGGCCUU-GCCUUUUCAAGUUCCCAACAUGG-AAGAGACAACAAAUUAAUGUUGUCGAAACACGAGAAAGACUUGUAAACUGUCGGCCGCUGAGG ..((((((((((...-))))((((...((((......))-))..(((((((......))))))))))).(((((.....)))))........))))))..... ( -30.20, z-score = -2.12, R) >droEre2.scaffold_4845 19478743 103 - 22589142 AUGCGGCUAGGUUUUGGCCCUUUCGCGUUCCCAACAUUGCAGGAGACAACAAAUUAAUGUUGUCGAAACACGAGAAAGACUUGUAAACUGUCGGCCGCUGAGG ..((((((.(((((.((..(((((.((((((..........)))(((((((......))))))).....))).))))).))...)))))...))))))..... ( -32.30, z-score = -1.78, R) >droYak2.chr2R 2870743 101 - 21139217 AUGCGGCUAGGCUCU-GCCCUUUCCAGUUCCCAACAUGG-AAGAGACAACAAAUUAAUGUUGUCGAAACACGAGAAAGACUUGUAAACUGUCGGCCGCUGAGG ..((((((.(((((.-....((((((((.....)).)))-))).(((((((......)))))))))...(((((.....))))).....)))))))))..... ( -30.30, z-score = -2.01, R) >droSec1.super_7 59604 101 - 3727775 UUGCGGCUAGGCCUU-GCCUCUGCAAGUUCCCAACAUGG-AAGAGACAACAAAUUAAUGUUGUCGAAACACGAGAAAGACUUGUAAACUGUCGGCCGCUGAGG ..((((((((((...-)))).((((((((.((.....))-....(((((((......))))))).............)))))))).......))))))..... ( -32.00, z-score = -2.40, R) >droSim1.chr2L 16125115 101 - 22036055 UUGCGGCUAGACCUU-GCCUCUGCAAGUUCCCAACAUGG-AAGAGACAACAAAUUAAUGUUGUCGAAACACGAGAAAGACUUGUAAACUGUCGGCCGUUGAGG ..((((((.((((((-((....)))))((((......))-))..(((((((......))))))).....(((((.....))))).....)))))))))..... ( -29.90, z-score = -2.35, R) >consensus UUGCGGCUAGGCCUU_GCCUUUUCAAGUUCCCAACAUGG_AAGAGACAACAAAUUAAUGUUGUCGAAACACGAGAAAGACUUGUAAACUGUCGGCCGCUGAGG ..((((((.(((....)))......((((...............(((((((......))))))).....(((((.....))))).))))...))))))..... (-24.90 = -24.98 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:08 2011