
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,598,065 – 1,598,155 |
| Length | 90 |
| Max. P | 0.817738 |

| Location | 1,598,065 – 1,598,155 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.80 |
| Shannon entropy | 0.42556 |
| G+C content | 0.35686 |
| Mean single sequence MFE | -12.76 |
| Consensus MFE | -7.50 |
| Energy contribution | -7.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.817738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1598065 90 + 23011544 ------GAGCCAGAGAAUACAAAAAACAAACUCACUUUUU----C-----CAACCUC-AGAAAUUCUAAUCCACUUAAAUGCUUAACAGCAUUAUGUGACCUUAAG- ------(((...(((((...................))))----)-----....)))-.............(((...((((((....))))))..)))........- ( -10.11, z-score = -1.46, R) >droPer1.super_8 1403669 91 + 3966273 ----------------ACACAGAGAAAAAUCUCCCCUUUUGGGCCAGAAACCACCUCGAAAAAUUCUAAUCCACUUAAAUGCUUAACAGCAUUAUGUGAGCCAAAAA ----------------.(((((((.....(((.(((....)))..)))......)))....................((((((....)))))).))))......... ( -15.40, z-score = -1.81, R) >dp4.chr4_group3 10198861 91 + 11692001 ----------------ACACAGAGAAAAAUCUGCCCUUUUGGGCCAGAAACCACCUCGAAAAAUUCUAAUCCACUUAAAUGCUUAACAGCAUUAUGUGACCCAAAAA ----------------.(((((((.....(((((((....))).))))......)))....................((((((....)))))).))))......... ( -20.40, z-score = -3.95, R) >droAna3.scaffold_12916 14998148 101 + 16180835 AAAAAAAGAAGGGAGAAGAGCAAAAACAAACUCACUUUUU----UAUAGUCAACCUC-AAAAAUUCUAAUCCACUUAAAUGCUUAACAGCAUUAUGUGACCUUAAG- ........(((((((..(((..........)))(((....----...)))....)))-.............(((...((((((....))))))..))).))))...- ( -14.70, z-score = -1.08, R) >droEre2.scaffold_4929 1644272 89 + 26641161 -------GAGCACAGAACACAAAAAACAAACUCACUUUUU----C-----CAACCUC-AGAAAUUCUAAUCCACUUAAAUGCUUAACAGCAUUAUGUGACCUUAAG- -------..........((((................(((----(-----.......-.))))..............((((((....)))))).))))........- ( -8.50, z-score = -1.21, R) >droYak2.chr2L 1571242 97 + 22324452 GAGCACUGAACACUGAACACAAAAAACAAACUCACUUUUU----CG----GAACCUC-AGAAAUUCUAAUCCACUUAAAUGCUUAACAGCAUUAUGUGACCUUAAG- (((..(((((...(((...............)))....))----))----)...)))-.............(((...((((((....))))))..)))........- ( -12.16, z-score = -1.44, R) >droSec1.super_14 1541144 93 + 2068291 ---GAGCGAGCAGAGAACACAAAAAACAAACUCACUUUUU----C-----CAACCUC-AGAAAUUCUAAUCCACUUAAAUGCUUAACAGCAUUAUGUGACCUUAAG- ---.........(((..((((................(((----(-----.......-.))))..............((((((....)))))).))))..)))...- ( -9.60, z-score = -0.35, R) >droSim1.chr2L 1571475 89 + 22036055 -------GAGCACGGAACACAAAAAACAAACUCACUUUUU----C-----CAACCUC-AGAAAUUCUAAUCCACUUAAAUGCUUAACAGCAUUAUGUGACCUUAAG- -------(((...((((.....................))----)-----)...)))-.............(((...((((((....))))))..)))........- ( -11.20, z-score = -2.09, R) >consensus _______GAGCACAGAACACAAAAAACAAACUCACUUUUU____C_____CAACCUC_AGAAAUUCUAAUCCACUUAAAUGCUUAACAGCAUUAUGUGACCUUAAG_ .......................................................................(((...((((((....))))))..)))......... ( -7.50 = -7.50 + 0.00)

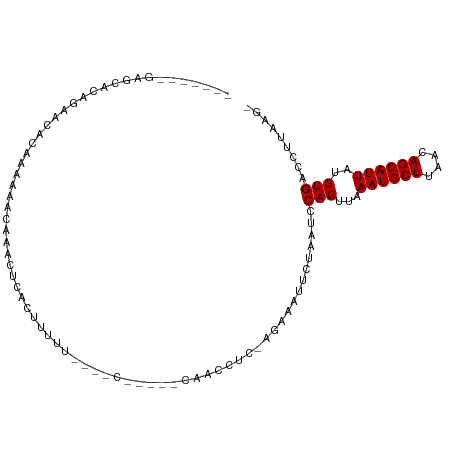
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:44 2011