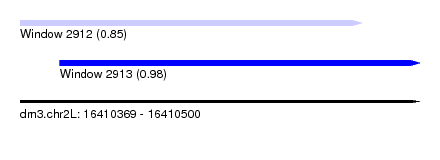
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,410,369 – 16,410,500 |
| Length | 131 |
| Max. P | 0.980583 |

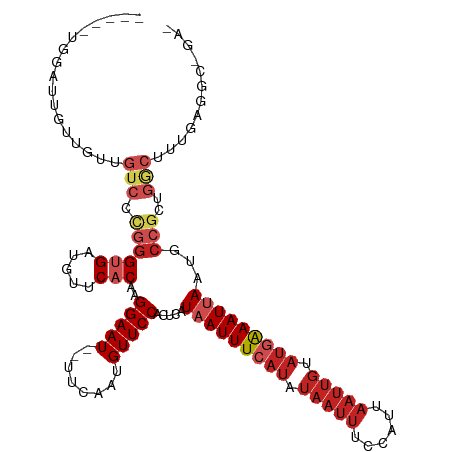
| Location | 16,410,369 – 16,410,481 |
|---|---|
| Length | 112 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.78 |
| Shannon entropy | 0.39817 |
| G+C content | 0.37630 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -13.72 |
| Energy contribution | -15.10 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16410369 112 + 23011544 -----UGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAG -----((((..((((..((.((..(((.....)))..)).))--..))))..))))(((.(((((((((((((((......)))))))))))))))........((((....))))))) ( -36.30, z-score = -3.75, R) >droSim1.chr2L 16121585 112 + 22036055 -----UGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAG -----((((..((((..((.((..(((.....)))..)).))--..))))..))))(((.(((((((((((((((......)))))))))))))))........((((....))))))) ( -36.30, z-score = -3.75, R) >droSec1.super_7 56197 112 + 3727775 -----UGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAG -----((((..((((..((.((..(((.....)))..)).))--..))))..))))(((.(((((((((((((((......)))))))))))))))........((((....))))))) ( -36.30, z-score = -3.75, R) >droYak2.chr2R 2867073 112 + 21139217 -----UGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAGGCCGCUGGCUUUGAGGCCGAG -----((((..((((..((.((..(((.....)))..)).))--..))))..))))(((.(((((((((((((((......))))))))))))))).((((.(.......).))))))) ( -38.90, z-score = -4.26, R) >droEre2.scaffold_4845 19475184 112 + 22589142 -----UGGAUUGUUGUCGUCCCUGGCGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAG -----((((..(((((((....)))))))..)).)).(((((--......))))).(((.(((((((((((((((......)))))))))))))))........((((....))))))) ( -33.10, z-score = -3.02, R) >droAna3.scaffold_12916 40139 112 - 16180835 -----UGGUUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGACAGAC -----(((..(((((..((.((..(((.....)))..)).))--..)))))..)))(((((((((((((((((((......)))))))))))))))..(((...)))..))))...... ( -33.70, z-score = -3.36, R) >dp4.chr4_group3 8131480 89 + 11692001 --------UGGACUGUUGUCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGCAUGAAAUGAAUG-------------------- --------.((((....))))...(((.....)))..(((((--......)))))...(((.(((((((.(((((......))))).)))))))..)))-------------------- ( -19.80, z-score = -0.84, R) >droPer1.super_1 9583077 93 + 10282868 --------UGGACUGUUGUCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGCAUGAAAUGAAUGCCCC---------------- --------.((((....)))).((((...((((....(((((--......))))).......(((((((.(((((......))))).))))))))))))))).---------------- ( -24.80, z-score = -2.13, R) >droWil1.scaffold_180772 4333745 101 - 8906247 -----AGUAUGAUUGUUGUCCUGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUGUAAUUUUCAUUAAUUGUAUGAAAUUAAUGCCGUUGGCU----------- -----......((((...((((..(((.....))).))))..--..))))..........(((((((((((((((......)))))))))))))))..(((...))).----------- ( -23.00, z-score = -1.48, R) >droVir3.scaffold_12963 13075351 103 + 20206255 UCGGUUUUGGCUCGGUUGUCCUGGGCGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUC----------CAUUAAUUGCAUAGAAUUAAUGCCGUCGACUCCACGAA---- (((......((((((.....))))))((((.......(((((--......)))))............----------((((((((......)))))))).)))).......))).---- ( -23.00, z-score = -0.13, R) >droGri2.scaffold_14978 511312 112 - 1124632 UCGGUUCUACAUCGGUUGUCCUGGGUGAUGUUCACAAGGAAUGUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGCAUAAAAUUAAUGCCGUUGACUUCGA------- ...........((((..(((.((((((.....)))..(((((((....)))))))......................((((((((......)))))))))))..))).))))------- ( -20.10, z-score = 0.10, R) >consensus _____UGGAUUGUUGUUGUCCCGGGUGAUGUUCACAAGGAAU__UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGC_GA_ .................(((...(((...........(((((........))))).....(((((((((.(((((......))))).)))))))))..)))...)))............ (-13.72 = -15.10 + 1.38)

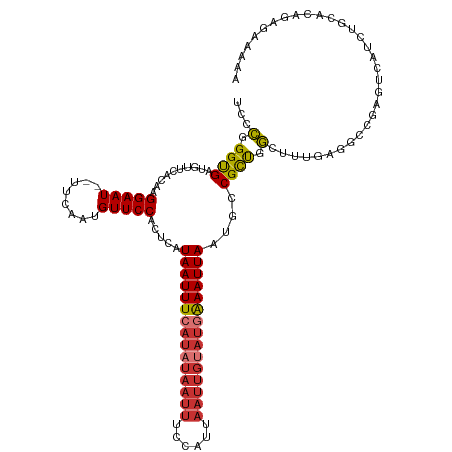
| Location | 16,410,382 – 16,410,500 |
|---|---|
| Length | 118 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.94 |
| Shannon entropy | 0.34732 |
| G+C content | 0.37806 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -18.43 |
| Energy contribution | -18.84 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16410382 118 + 23011544 UCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAGUCAUCUGCACAGAGAAAAA ...(((((((.........(((((--......))))).(((.(((((((((((((((......)))))))))))))))........((((....))))))))))))))............ ( -34.40, z-score = -3.13, R) >droSim1.chr2L 16121598 118 + 22036055 UCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAGUCAUCUGCACAGAGAAAAA ...(((((((.........(((((--......))))).(((.(((((((((((((((......)))))))))))))))........((((....))))))))))))))............ ( -34.40, z-score = -3.13, R) >droSec1.super_7 56210 118 + 3727775 UCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAGUCUUCUGCACAGAGAAAAA ..((..(((.....)))..))..(--(((..(((...((((.(((((((((((((((......)))))))))))))))........((((....)))))))).....)))..)))).... ( -32.80, z-score = -2.67, R) >droYak2.chr2R 2867086 118 + 21139217 UCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAGGCCGCUGGCUUUGAGGCCGAGUCAUCUGCACAGAGAAAAA ...(((((((.........(((((--......))))).(((.(((((((((((((((......))))))))))))))).((((.(.......).))))))))))))))............ ( -37.00, z-score = -3.69, R) >droEre2.scaffold_4845 19475197 118 + 22589142 UCCCUGGCGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAGUCAUCUGCACAGCGAAAAA ((.(((((((((.......(((((--......)))))((((.(((((((((((((((......)))))))))))))))........((((....)))))))))))).)).))).)).... ( -37.70, z-score = -4.48, R) >droAna3.scaffold_12916 40152 118 - 16180835 UCCCGGGUGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGACAGACUCUUCUGCACAGAACAAAA ..((..(((.....)))..))...--.....(((((..(((((((((((((((((((......)))))))))))))))..(((...)))..)))).((((....))))....)))))... ( -32.30, z-score = -3.49, R) >droVir3.scaffold_12963 13075369 91 + 20206255 UCCUGGGCGAUGUUCACAAGGAAU--UUCAAUGUUCCACUCAUAAUUUC----------CAUUAAUUGCAUAGAAUUAAUGCCGUCGACUCCACGAAUUGAGC----------------- ((.(((((((((.......(((((--......)))))............----------((((((((......)))))))).))))).).))).)).......----------------- ( -19.60, z-score = -1.10, R) >droGri2.scaffold_14978 511330 94 - 1124632 UCCUGGGUGAUGUUCACAAGGAAUGUUUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGCAUAAAAUUAAUGCCGUUGACUUCGA-------------------------- ...((((((.....)))..(((((((....))))))).....................)))(((((.((((.......)))).)))))......-------------------------- ( -15.60, z-score = -0.18, R) >consensus UCCCGGGUGAUGUUCACAAGGAAU__UUCAAUGUUCCACUCAUAAUUUCAUAUAAUUUCCAUUAAUUGUAUGAAAUUAAUGCCGCUGGCUUUGAGGCCGAGUCAUCUGCACAGAGAAAAA ...(.((((..........(((((........))))).....(((((((((((((((......)))))))))))))))....)))).)................................ (-18.43 = -18.84 + 0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:06 2011