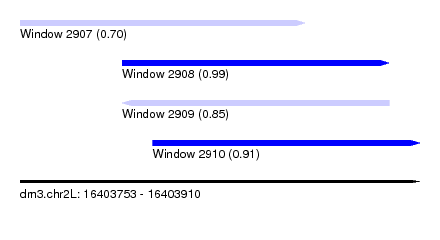
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,403,753 – 16,403,910 |
| Length | 157 |
| Max. P | 0.988878 |

| Location | 16,403,753 – 16,403,865 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.28 |
| Shannon entropy | 0.40316 |
| G+C content | 0.42877 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -20.23 |
| Energy contribution | -21.28 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.703178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
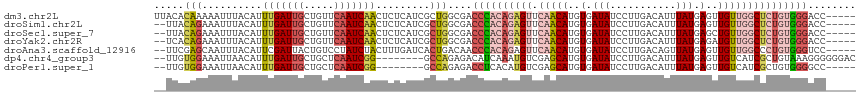
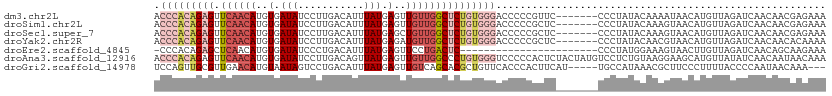
>dm3.chr2L 16403753 112 + 23011544 UUACACAAAAAUUUACAUUUGAUUGCUGUUCAAUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACC----- ..................(((((((.....)))))))......(((....)))((((((((((.(((((..(.(((...........))).)..)))))))))))))))...----- ( -30.50, z-score = -2.16, R) >droSim1.chr2L 16114213 110 + 22036055 --UUACAGAAAUUUACAUUUGAUUGCUGUUCAAUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACC----- --...(((..........(((((((.....))))))).........)))....((((((((((.(((((..(.(((...........))).)..)))))))))))))))...----- ( -31.11, z-score = -2.10, R) >droSec1.super_7 49629 110 + 3727775 --UUACAGAAAUUUACAUUUGAUUGCUGUUCAAUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGCUGUUGGCUCUGUGGGACC----- --...(((..........(((((((.....))))))).........)))....((((((((((.(((((.((.(((...........)))..)))))))))))))))))...----- ( -31.21, z-score = -2.17, R) >droYak2.chr2R 2860415 110 + 21139217 --UCACAGAAAUUUACAUUUGAUUGCUGUUCAAUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGAUGUUGGCUCUGUGGGACC----- --...(((..........(((((((.....))))))).........)))....((((((((((.((((((.(.(((...........))).).))))))))))))))))...----- ( -34.61, z-score = -2.91, R) >droAna3.scaffold_12916 33143 110 - 16180835 --UUCGAGCAAUUUACAUUCGAUUACUGUCCUAUCUACUUUGAUCACUGACAACCCACAGAGUUCAACAUGUGAUAUCCUUGACAGUUAUGAGUUGUUGGCCCUGUGGGUCC----- --.(((((.........)))))....((((..(((......)))....))))((((((((.(..(((((..(.(((...........))).)..)))))..)))))))))..----- ( -26.10, z-score = -0.78, R) >dp4.chr4_group3 8125810 107 + 11692001 --UUGUGGAAAUUAACAUUUGAUUGCUGCUCAAUCGG--------GCCAGAGACAUCAAAUGUCGAGCAUGUGAUAUCCUUGACAUUUAUGAGUUGUCAUCGCUGUAAAGGGGGGAC --..(((....(..(((((((((..((((((....))--------).)))....)))))))))..).)))......(((((((((.........))))).(.((....)).))))). ( -23.30, z-score = 0.85, R) >droPer1.super_1 9577391 102 + 10282868 --UUGUGGAAAUUAACAUUUGAUUGCUGCUCAAUCGG--------GCCAGAGACCUCACAUGUCGAGCAUGUGAUAUCCUUGACAUUUAUGAGUUGUCAUCGCUGUGGGGCC----- --.(((........)))..((((((.....))))))(--------(((((.((..(((((((.....)))))))..))))(((((.........))))).........))))----- ( -24.90, z-score = 0.61, R) >consensus __UUACAGAAAUUUACAUUUGAUUGCUGUUCAAUCAACUCUCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACC_____ .....(((..........(((((((.....))))))).........)))....((((((((((.(((((..(.(((...........))).)..)))))))))))))))........ (-20.23 = -21.28 + 1.05)
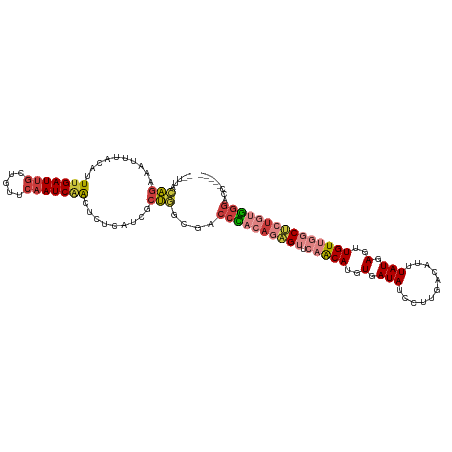
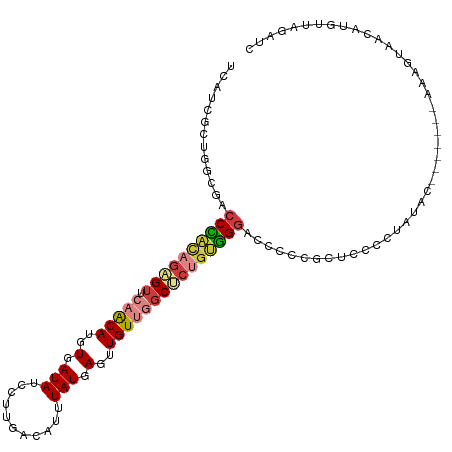
| Location | 16,403,793 – 16,403,898 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.40 |
| Shannon entropy | 0.59978 |
| G+C content | 0.48027 |
| Mean single sequence MFE | -27.11 |
| Consensus MFE | -16.28 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16403793 105 + 23011544 UCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACCCCCGUUCCCCUAUAC-------AAAAUAACAUGUUAGAUC ......((((((.((((((((((.(((((..(.(((...........))).)..)))))))))))))))......((........))-------.........))))))... ( -28.20, z-score = -1.76, R) >droSim1.chr2L 16114251 105 + 22036055 UCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACCCCCGCUCCCCUAUAC-------AAAGUAACAUGUUAGAUC .....((.((.(.((((((((((.(((((..(.(((...........))).)..))))))))))))))).).)).))....(((.((-------(........))))))... ( -29.80, z-score = -1.97, R) >droSec1.super_7 49667 105 + 3727775 UCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGCUGUUGGCUCUGUGGGACCCCCGCUCCCCUAUAC-------AAAGUAACAUGUUAGAUC .....((.((.(.((((((((((.(((((.((.(((...........)))..))))))))))))))))).).)).))....(((.((-------(........))))))... ( -29.90, z-score = -1.95, R) >droYak2.chr2R 2860453 105 + 21139217 UCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGAUGUUGGCUCUGUGGGACCCCCGCUCCCCUAUAC-------AACGUAACAUGUUAGAUC .....((.((.(.((((((((((.((((((.(.(((...........))).).)))))))))))))))).).)).))....(((.((-------(........))))))... ( -33.30, z-score = -3.02, R) >droAna3.scaffold_12916 33181 112 - 16180835 UGAUCACUGACAACCCACAGAGUUCAACAUGUGAUAUCCUUGACAGUUAUGAGUUGUUGGCCCUGUGGGUCCCCCACUCUACUAUGUCCUCUGUAAGGAAGCAUGUUAUAUC .((.(.(((........))).).))(((((((....(((((((((.....((((.(..(((((...)))))..).)))).....))))......))))).)))))))..... ( -28.10, z-score = -0.26, R) >droPer1.super_1 9577426 82 + 10282868 -----GCCAGAGACCUCACAUGUCGAGCAUGUGAUAUCCUUGACAUUUAUGAGUUGUCAUCGCUGUGGGGCCCCCUCCCCCCUCGCC------------------------- -----(((((.((..(((((((.....)))))))......(((((.........))))))).))).((((........))))..)).------------------------- ( -22.80, z-score = -1.11, R) >droGri2.scaffold_14978 605963 105 + 1124632 --AAUUUGAACAUCCAGUUGCGUUGAACAUGUAAUAGUCCUGACAUUUAUGAGUUGUCAGCAC-GCUGUUCACCCACUUCAUUGCCA----UAAACGCUUCCCUUUUACCCC --....((((((....(((((((.....))))))).((.((((((.........)))))).))-..))))))...............----..................... ( -17.70, z-score = -1.11, R) >consensus UCAUCGCUGGCGACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACCCCCGCUCCCCUAUAC_______AAAGUAACAUGUUAGAUC .............((((((((((.(((((..(.(((...........))).)..)))))))))))))))........................................... (-16.28 = -17.20 + 0.92)

| Location | 16,403,793 – 16,403,898 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.40 |
| Shannon entropy | 0.59978 |
| G+C content | 0.48027 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -16.22 |
| Energy contribution | -16.86 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16403793 105 - 23011544 GAUCUAACAUGUUAUUUU-------GUAUAGGGGAACGGGGGUCCCACAGAGCCAACAACUCAUAAAUGUCAAGGAUAUCACAUGUUGAACUCUGUGGGUCGCCAGCGAUGA ..(((((((........)-------)).))))......((.(.(((((((((....((((......(((((...))))).....))))..))))))))).).))........ ( -28.70, z-score = -0.63, R) >droSim1.chr2L 16114251 105 - 22036055 GAUCUAACAUGUUACUUU-------GUAUAGGGGAGCGGGGGUCCCACAGAGCCAACAACUCAUAAAUGUCAAGGAUAUCACAUGUUGAACUCUGUGGGUCGCCAGCGAUGA ..(((((((........)-------)).))))...((.((.(.(((((((((....((((......(((((...))))).....))))..))))))))).).)).))..... ( -31.90, z-score = -1.26, R) >droSec1.super_7 49667 105 - 3727775 GAUCUAACAUGUUACUUU-------GUAUAGGGGAGCGGGGGUCCCACAGAGCCAACAGCUCAUAAAUGUCAAGGAUAUCACAUGUUGAACUCUGUGGGUCGCCAGCGAUGA ..(((((((........)-------)).))))...((.((.(.(((((((((.(((((........(((((...)))))....)))))..))))))))).).)).))..... ( -32.80, z-score = -1.27, R) >droYak2.chr2R 2860453 105 - 21139217 GAUCUAACAUGUUACGUU-------GUAUAGGGGAGCGGGGGUCCCACAGAGCCAACAUCUCAUAAAUGUCAAGGAUAUCACAUGUUGAACUCUGUGGGUCGCCAGCGAUGA ..(((((((........)-------)).))))...((.((.(.(((((((((.((((((.......(((((...)))))...))))))..))))))))).).)).))..... ( -33.00, z-score = -1.31, R) >droAna3.scaffold_12916 33181 112 + 16180835 GAUAUAACAUGCUUCCUUACAGAGGACAUAGUAGAGUGGGGGACCCACAGGGCCAACAACUCAUAACUGUCAAGGAUAUCACAUGUUGAACUCUGUGGGUUGUCAGUGAUCA (((...((.(((((((((...)))))...)))).....((.(((((((((((.(((((....(((.((.....)).)))....)))))..))))))))))).)).)).))). ( -33.40, z-score = -0.81, R) >droPer1.super_1 9577426 82 - 10282868 -------------------------GGCGAGGGGGGAGGGGGCCCCACAGCGAUGACAACUCAUAAAUGUCAAGGAUAUCACAUGCUCGACAUGUGAGGUCUCUGGC----- -------------------------.((...((((.......))))...)).((((....))))....((((..(((.(((((((.....))))))).)))..))))----- ( -28.00, z-score = -1.37, R) >droGri2.scaffold_14978 605963 105 - 1124632 GGGGUAAAAGGGAAGCGUUUA----UGGCAAUGAAGUGGGUGAACAGC-GUGCUGACAACUCAUAAAUGUCAGGACUAUUACAUGUUCAACGCAACUGGAUGUUCAAAUU-- .(..((...((...(((((..----(((((.(...((((((...(((.-...)))...)))))).).)))))((((........)))))))))..))...))..).....-- ( -24.70, z-score = -0.89, R) >consensus GAUCUAACAUGUUACUUU_______GUAUAGGGGAGCGGGGGUCCCACAGAGCCAACAACUCAUAAAUGUCAAGGAUAUCACAUGUUGAACUCUGUGGGUCGCCAGCGAUGA ......................................((.(.(((((((((....((((......(((((...))))).....))))..))))))))).).))........ (-16.22 = -16.86 + 0.64)

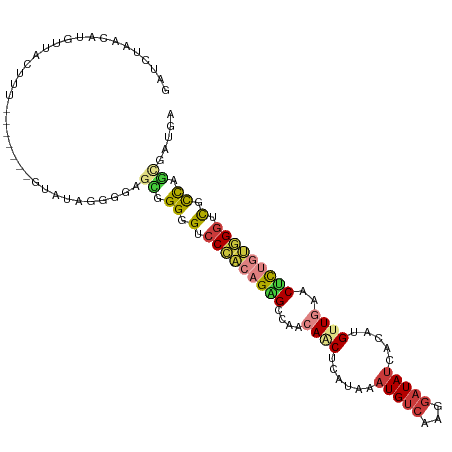
| Location | 16,403,805 – 16,403,910 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Shannon entropy | 0.49974 |
| G+C content | 0.43071 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -14.23 |
| Energy contribution | -16.09 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16403805 105 + 23011544 ACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACCCCCGUUC-------CCCUAUACAAAAUAACAUGUUAGAUCAACAACGAGAAA .((((((((((.(((((..(.(((...........))).)..)))))))))))))))......(((.-------..(((.(((........))))))...)))......... ( -25.90, z-score = -1.91, R) >droSim1.chr2L 16114263 105 + 22036055 ACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACCCCCGCUC-------CCCUAUACAAAGUAACAUGUUAGAUCAACAACGAGAAA .((((((((((.(((((..(.(((...........))).)..))))))))))))))).......(((-------.....(((...)))...((((.....))))..)))... ( -25.20, z-score = -1.43, R) >droSec1.super_7 49679 105 + 3727775 ACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGCUGUUGGCUCUGUGGGACCCCCGCUC-------CCCUAUACAAAGUAACAUGUUAGAUCAACAACGAGAAA .((((((((((.(((((.((.(((...........)))..))))))))))))))))).......(((-------.....(((...)))...((((.....))))..)))... ( -25.30, z-score = -1.53, R) >droYak2.chr2R 2860465 105 + 21139217 ACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGAUGUUGGCUCUGUGGGACCCCCGCUC-------CCCUAUACAACGUAACAUGUUAGAUCAACAACACAAAA .((((((((((.((((((.(.(((...........))).).))))))))))))))))..........-------..(((.(((........))))))............... ( -28.40, z-score = -2.77, R) >droEre2.scaffold_4845 19469191 88 + 22589142 -CCCACAGAGCUCAACAUGUGAUAUCCCUGACAUUUAUGAGUUCCUGACUC-----------------------CCCUAUGGAAAGUAACUUGUUAGAUCAACAGCAAGAAA -....(((((((((..((((.(......).))))...)))))).)))..((-----------------------(.....)))......((((((........))))))... ( -16.90, z-score = -1.10, R) >droAna3.scaffold_12916 33193 112 - 16180835 ACCCACAGAGUUCAACAUGUGAUAUCCUUGACAGUUAUGAGUUGUUGGCCCUGUGGGUCCCCCACUCUACUAUGUCCUCUGUAAGGAAGCAUGUUAUAUCAACAAUAACAAA .......((....(((((((....(((((((((.....((((.(..(((((...)))))..).)))).....))))......))))).)))))))...))............ ( -26.50, z-score = -0.52, R) >droGri2.scaffold_14978 605973 104 + 1124632 UCCAGUUGCGUUGAACAUGUAAUAGUCCUGACAUUUAUGAGUUGUCAGCACGCUGUUCACCCACUUCAU-----UGCCAUAAACGCUUCCCUUUUACCCCAAUAACAAA--- .......((((((((((.((....((.((((((.........)))))).)))))))))...((......-----)).....))))).......................--- ( -16.30, z-score = -0.91, R) >consensus ACCCACAGAGUUCAACAUGUGAUAUCCUUGACAUUUAUGAGUUGUUGGCUCUGUGGGACCCCCGCUC_______CCCUAUACAAAGUAACAUGUUAGAUCAACAACAAGAAA .(((((((((.((((((..(.(((...........))).)..)))))))))))))))....................................................... (-14.23 = -16.09 + 1.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:03 2011