
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,390,497 – 16,390,591 |
| Length | 94 |
| Max. P | 0.854694 |

| Location | 16,390,497 – 16,390,591 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.88 |
| Shannon entropy | 0.65870 |
| G+C content | 0.50894 |
| Mean single sequence MFE | -19.50 |
| Consensus MFE | -9.37 |
| Energy contribution | -9.21 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
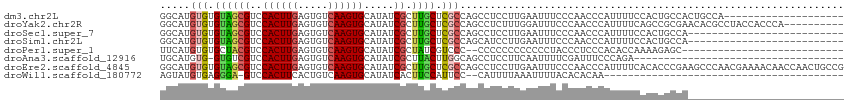
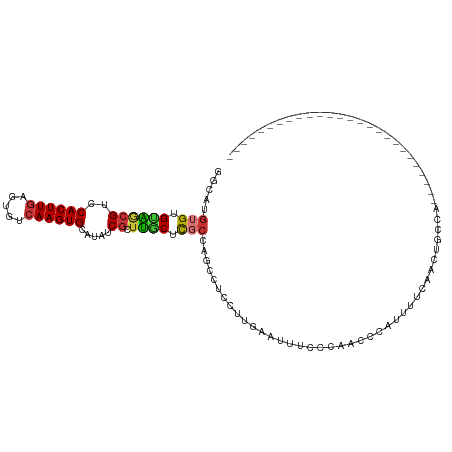
>dm3.chr2L 16390497 94 + 23011544 GGCAUGUGUGUAGCGUCCACUUGAGUGUCAAGUGCAUAUCGCUUGCUCGCCAGCCUCCUUGAAUUUCCCAACCCAUUUUCCACUGCCACUGCCA-------------------- ((((.(((.((((.(..((((((.....))))))......(((.(....).)))..........................).))))))))))).-------------------- ( -23.80, z-score = -1.55, R) >droYak2.chr2R 2846854 104 + 21139217 GGCAUGUGUGUAGCGUCCACUUGAGUGUCAAGUGCAUAUCGCUUGCUCGCCAGCCUCUUUGGAUUUCCCAACCCAUUUUCAGCCGCGAACACGCCUACCACCCA---------- ((...((((((((((..((((((.....)))))).....))))...((((..((.....(((..........)))......)).))))))))))...)).....---------- ( -27.00, z-score = -1.27, R) >droSec1.super_7 35453 88 + 3727775 GGCAUGUGUGUAGCGUCCACUUGAGUGUCAAGUGCAUAUCGCUUGCUCGCCAGCCUCCUUGAAUUUCCCAACCCAUUUUCCACUGCCA-------------------------- ((((.(((...((((..((((((.....)))))).....)))).(((....)))..........................))))))).-------------------------- ( -20.80, z-score = -1.32, R) >droSim1.chr2L 16099404 88 + 22036055 GGCAUGUGUGUAGCGUCCACUUGAGUGUCAAGUGCAUAUCGCUUGCUCGCCAGCAUCCUUGAAUUUCCCAACCCAUUUUCCACUGCCA-------------------------- ((((.(((...((((..((((((.....)))))).....))))((((....)))).........................))))))).-------------------------- ( -22.40, z-score = -1.67, R) >droPer1.super_1 9565165 85 + 10282868 UUCAUGUGUGCUACGUCCACUUGAGUGUCAAGUGCAUAUCGCUAUCGUCCC--CCCCCCCCCCCCUACCCUCCCACACCAAAAGAGC--------------------------- .....(((((....((.((((((.....)))))).))...((....))...--....................))))).........--------------------------- ( -10.40, z-score = -0.93, R) >droAna3.scaffold_12916 21475 78 - 16180835 UGCAUGUG-GUGUCGUCCACUUGAGUGUCAAGUGCAUAUCGCUUACUUGGCAGCCUCCUUCAAUUUUCGAUUUCCCAGA----------------------------------- ....((.(-(.((((.....(((((((((((((((.....))..))))))))......)))))....))))..))))..----------------------------------- ( -16.80, z-score = -0.27, R) >droEre2.scaffold_4845 19456970 114 + 22589142 GGCAUGUGUGUAGCGUCCACUUGAGUGUCAAGUGCAUAUCGCUUGCUCGCCAGCCUCCUUGAAUUUCCCAACCCAUUUUCACACCCGAAGCCCAACGAAAACAACCAACUGCCG ((((.(((.((((((..((((((.....)))))).....))).))).)))..((.((..((((..............)))).....)).))..................)))). ( -22.54, z-score = -0.49, R) >droWil1.scaffold_180772 4527817 71 + 8906247 AGUAUGUGAGGGA-GUCCACUUCACUGUCAAGUGCAUAUCACUUCCAUUCC--CAUUUUAAAUUUUACACACAA---------------------------------------- .((.(((((((((-((......((((....))))......)))))).....--...........))))).))..---------------------------------------- ( -12.29, z-score = -0.97, R) >consensus GGCAUGUGUGUAGCGUCCACUUGAGUGUCAAGUGCAUAUCGCUUGCUCGCCAGCCUCCUUGAAUUUCCCAACCCAUUUUCAACUGCCA__________________________ .....(((.((((((..((((((.....)))))).....)).)))).)))................................................................ ( -9.37 = -9.21 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:59 2011