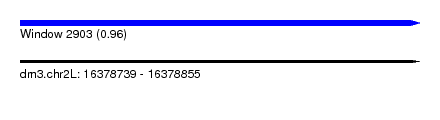
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,378,739 – 16,378,855 |
| Length | 116 |
| Max. P | 0.960587 |

| Location | 16,378,739 – 16,378,855 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.09 |
| Shannon entropy | 0.48688 |
| G+C content | 0.43537 |
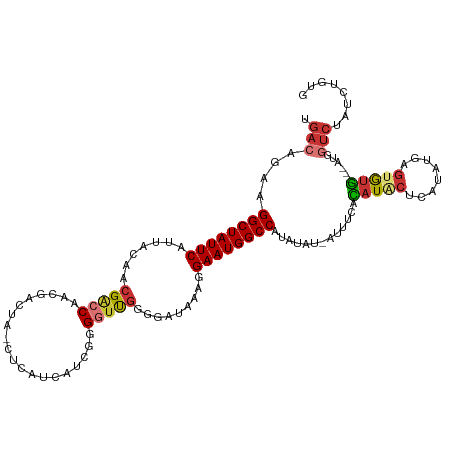
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -15.65 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
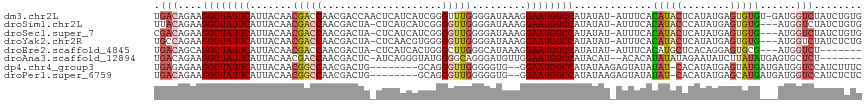
>dm3.chr2L 16378739 116 + 23011544 UGACAGAAGGCUAUUCAUUACAACGACCAACGACCAACUCAUCAUCGGGUUUGGGGAUAAAGGAAUGGCCAUAUAU-AUUUCACAUACUCAUAUGAGUGUGU-GAUGGUCUAUCUGUG ..(((((.((((((((.......((.....)).(((((((......))).))))........))))))))......-...((((((((((....))))))))-)).......))))). ( -38.70, z-score = -3.59, R) >droSim1.chr2L 16087742 113 + 22036055 UUACAGAAGGCUAUUCAUUACAACGACCAACGACUA-CUCAUCAUCGGGGUUGGGGAUAAAGGAAUGGCCAUAUAU-AUUUCACAUACCCAUAUGAGUGUG---AUGGUCUAUCUGUG .((((((.((((((((.((((..(((((..(((...-.......))).)))))..).)))..))))))))......-...((((((((.(....).)))))---.)))....)))))) ( -31.20, z-score = -1.68, R) >droSec1.super_7 23867 113 + 3727775 CGACAGAAGGCUAUUCAUUACAACGACCAACGACUA-CUCAUCAUCGGGGUUGGGGAUAAAGGAAUGGCCAUAUAU-AUUUCACAUACUCAUAUGAGUGUG---AUGGUCUAUCUGUG ..(((((.((((((((.((((..(((((..(((...-.......))).)))))..).)))..))))))))......-...((((((((((....)))))))---.)))....))))). ( -35.90, z-score = -3.44, R) >droYak2.chr2R 2833916 113 + 21139217 UGCCAGAAGGCUAUUCAUUACAACGACCAACGACUA-CUCAACGUGGGGGUUGGGGAUAAAGGAAUGGCCAUAUAU-AUUUCACAUACUCAUAUGAGUGUG---AUGGUCUAUCUCUG ...((((.((((((((..........(((((..(((-(.....))))..)))))........))))))))....((-(..((((((((((....)))))))---.)))..))).)))) ( -37.57, z-score = -3.50, R) >droEre2.scaffold_4845 19445590 106 + 22589142 UGACAGCAGGCUAUUCAUUACAACGACCAACGACUA-CUCAUCACUGGGCUUGGGCAUAAAGGAAUGGCCAUAUAU-AUUUCACAUGCUCACAGGAGUGCG---AUGGUCU------- .(((.((.((((((((..........((((...(((-........)))..))))........))))))))......-.........((((....)))))).---...))).------- ( -24.67, z-score = -0.63, R) >droAna3.scaffold_12894 5195 108 - 7079 UGACAGAAGGCUAUUCAUUACAACGACCAACGACUC-AUCAGGGUAUGGGGCAGGGAUGUUGGAAUGGCCAUACAU--ACACAUAUAUAGAAUAUCUUAUAUGAGUGCUCU------- ....(((.((((((((....(((((.((.....(((-((......)))))....)).))))))))))))).....(--((.((((((.((.....)))))))).))).)))------- ( -27.80, z-score = -1.83, R) >dp4.chr4_group3 8089320 107 + 11692001 UGAGAGAAGGCUAUUCAUUACAACGGCCAACGACUG--------GCAGGGUUGGGGGUG--GGAAUGGCCAUAUAAGAGUAUAUAU-CACAUAUGAGUAUGAUGAUGGUCCAUCUUUC .((((((.((((((((.(..((((.((((.....))--------))...))))..)...--.)))))))).............(((-((((((....)))).))))).....)))))) ( -32.40, z-score = -2.23, R) >droPer1.super_6759 1347 107 + 1607 UGACAGAAGGCUAUUCAUUACAACGGCCAACGACUG--------GCAGGGUUGGGGGUG--GGAAUGGCCAUAUAAGAGUAUAUAU-CACAUAUGAGCAUGAUGAUGGUCCAUCUCUC ........((((((((.(..((((.((((.....))--------))...))))..)...--.)))))))).....((((....(((-(((((......))).)))))......)))). ( -29.70, z-score = -0.86, R) >consensus UGACAGAAGGCUAUUCAUUACAACGACCAACGACUA_CUCAUCAUCGGGGUUGGGGAUAAAGGAAUGGCCAUAUAU_AUUUCACAUACUCAUAUGAGUGUG___AUGGUCUAUCUGUG .(((....((((((((..........(((((..((...........)).)))))........)))))))).............(((((........)))))......)))........ (-15.65 = -16.50 + 0.85)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:58 2011