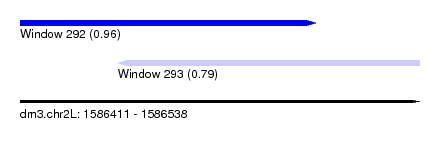
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,586,411 – 1,586,538 |
| Length | 127 |
| Max. P | 0.963945 |

| Location | 1,586,411 – 1,586,505 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 63.55 |
| Shannon entropy | 0.69912 |
| G+C content | 0.60416 |
| Mean single sequence MFE | -39.79 |
| Consensus MFE | -17.53 |
| Energy contribution | -21.09 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.963945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
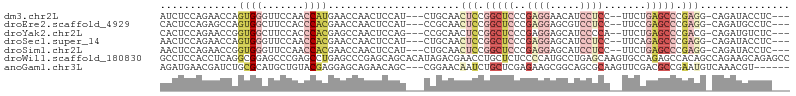
>dm3.chr2L 1586411 94 + 23011544 -------GGCCUCGGCAGGCACAUCCAUUGGUGCUUUUGAGGUAUCUGCCUCGGGCUCAGAAGGAGGAUGUUCCUCGGGAGCCGGAGUUGCAGAUGGAGUU----- -------.(((((((.((((((........)))))))))))))(((((((((.(((((.....((((.....))))..))))).)))..))))))......----- ( -44.00, z-score = -2.88, R) >droEre2.scaffold_4929 1632522 94 + 26641161 -------GGCCUCGGCCGGCACAUCCUUCUGAGCUUUUGAGGCAUCUGCCUCGGGCUCGGAAGGAGGACGCUCCUCGGGAGCCGGAGUUGCGGAUGGAGUU----- -------(((....))).(..(.((((((((((((..((((((....)))))))))))))))))))..)(((((((.(.(((....))).).)).))))).----- ( -47.90, z-score = -2.12, R) >droYak2.chr2L 1559525 88 + 22324452 -------GACCUCGGCAGG-AUGUU-----GUGCUUUCGAGACAUCUGCGUCGGGCUCAGAAUGGGGAUGCUCCUCGGGAGCCGGAGUUGCGGCUGGAGUU----- -------(((..((((.((-(((((-----.((....)).)))))))(((((.(((((....(((((.....))))).))))).))..))).))))..)))----- ( -35.50, z-score = -0.81, R) >droSec1.super_14 1529665 94 + 2068291 -------GGCCUCGGCAGGCACAUCCAUUGGUGCUUUUGAGGUAUCUGCCUCGGGCUCUGAAGGAGGAUGCUCCUCGGGAGCCGGAGUUGCAGAUGGAGUU----- -------.(((((((.((((((........)))))))))))))(((((((((.((((((..(((((....)))))..)))))).)))..))))))......----- ( -48.40, z-score = -3.61, R) >droSim1.chr2L 1559934 94 + 22036055 -------GGCCUCGGCAGGCACAUCCAUUGGUGCUUUUGAGGUAUCUGCCUCGGGCUCAGAAGGAGGAUGCUCCUCGGGAGCCGGAGUUGCAGAUGGAGUU----- -------.(((((((.((((((........)))))))))))))(((((((((.(((((...(((((....)))))...))))).)))..))))))......----- ( -46.40, z-score = -3.17, R) >droWil1.scaffold_180830 111097 96 + 119711 -------AUCCAAUUCUGGCUCCUUUUCGGGUGCAGCUGGC---UCUGCUUCUGGCUGUGGCUCUGGCACUUGCUCAGGCAUGGGGAGAGCAGGUUCGUCUAUGUG -------...........((.(((....))).))((((.((---(((.((..((.(((.(((..........)))))).))..)).))))).)))).......... ( -28.80, z-score = 1.58, R) >anoGam1.chr3L 16936493 94 + 41284009 GCCUGCCGCUCCUGCCGCGCACUGUC-CUGCAACGUUUGACAU-UCGGCGUCGAACUUG---CGCUGCCGCUUCUCGAGCAG-AUUGUUCCGGCUGUUCU------ (((.((.((....)).))(((.....-..((((.(((((((..-.....))))))))))---).((((((.....)).))))-..)))...)))......------ ( -27.50, z-score = 0.49, R) >consensus _______GGCCUCGGCAGGCACAUCCAUUGGUGCUUUUGAGGUAUCUGCCUCGGGCUCAGAAGGAGGAUGCUCCUCGGGAGCCGGAGUUGCAGAUGGAGUU_____ .........((((((.((((.(........).))))))))))((((((((((.(((((.....((((.....))))..))))).)))..))))))).......... (-17.53 = -21.09 + 3.56)


| Location | 1,586,442 – 1,586,538 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 63.41 |
| Shannon entropy | 0.70759 |
| G+C content | 0.60727 |
| Mean single sequence MFE | -32.19 |
| Consensus MFE | -6.94 |
| Energy contribution | -9.23 |
| Covariance contribution | 2.29 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.790220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1586442 96 - 23011544 AUCUCCAGAACCAGUGGGUUCCAACCAUGAACCAACUCCAU---CUGCAACUCCGGCUCCCGAGGAACAUCCUCC--UUCUGAGCCCGAGG-CAGAUACCUC--- ............(((.(((((.......))))).)))..((---((((..(((.(((((..(((((......)))--))..))))).))))-))))).....--- ( -33.00, z-score = -3.65, R) >droEre2.scaffold_4929 1632553 96 - 26641161 CACUCCAGAGCCAGUGGCUUCCACCCACGAACCAACUCCAU---CCGCAACUCCGGCUCCCGAGGAGCGUCCUCC--UUCCGAGCCCGAGG-CAGAUGCCUC--- .......(((...((((.......))))..........(((---(.((..(((.(((((..((((((....))))--))..))))).))))-).)))).)))--- ( -31.00, z-score = -2.02, R) >droYak2.chr2L 1559550 96 - 22324452 CACUCCAGAACCGGUGGCUUCCACCCACGAGCCAACUCCAG---CCGCAACUCCGGCUCCCGAGGAGCAUCCCCA--UUCUGAGCCCGACG-CAGAUGUCUC--- ..(((.((((..((((((((........)))))).((((((---(((......))))).....))))....))..--)))))))...((((-....))))..--- ( -26.60, z-score = -0.83, R) >droSec1.super_14 1529696 96 - 2068291 AACUCCAGAACCAGUGGGUUCCAACCACGAACCAACUCCAU---CUGCAACUCCGGCUCCCGAGGAGCAUCCUCC--UUCAGAGCCCGAGG-CAGAUACCUC--- ............(((.(((((.......))))).)))..((---((((..(((.(((((..((((((....))))--))..))))).))))-))))).....--- ( -35.40, z-score = -4.33, R) >droSim1.chr2L 1559965 96 - 22036055 AACUCCAGAACCGGUGGGUUCCAACCACGAACCAACUCCAU---CUGCAACUCCGGCUCCCGAGGAGCAUCCUCC--UUCUGAGCCCGAGG-CAGAUACCUC--- ............(((((((((.......))))).......(---((((..(((.(((((..((((((....))))--))..))))).))))-))))))))..--- ( -36.60, z-score = -3.99, R) >droWil1.scaffold_180830 111128 105 - 119711 GCCUCCACCUCAGGCGGAGCCCGAGCCUGAGCCCGAGCAGCACAUAGACGAACCUGCUCUCCCCAUGCCUGAGCAAGUGCCAGAGCCACAGCCAGAAGCAGAGCC (((((...(((((((.........)))))))...)))..))............(((((((......(((((.((....))))).)).......)).))))).... ( -30.02, z-score = -0.18, R) >anoGam1.chr3L 16936524 96 - 41284009 AGAUGAACGAUCUGCGCAUGCUGUACGAGGAGCAGAACAGC---CGGAACAAUCUGCUCGAGAAGCGGCAGCGCAAGUUCGACGCCGAAUGUCAAACGU------ ....((((....(((((.((((((.....(((((((.....---........))))))).....))))))))))).))))((((.....))))......------ ( -32.72, z-score = -1.11, R) >consensus AACUCCAGAACCAGUGGGUUCCAACCACGAACCAACUCCAU___CUGCAACUCCGGCUCCCGAGGAGCAUCCUCC__UUCUGAGCCCGAGG_CAGAUGCCUC___ .............((((.......))))......................(((.(((((..((((.....)))).......))))).)))............... ( -6.94 = -9.23 + 2.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:43 2011