| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,356,686 – 16,356,776 |
| Length | 90 |
| Max. P | 0.967061 |

| Location | 16,356,686 – 16,356,776 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 57.88 |
| Shannon entropy | 0.74100 |
| G+C content | 0.45211 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -8.47 |
| Energy contribution | -8.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
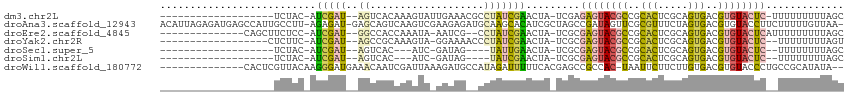
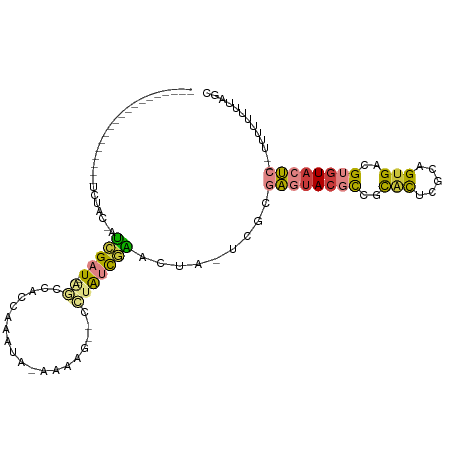
>dm3.chr2L 16356686 90 - 23011544 -------------------UCUAC-AUCGAU--AGUCACAAAGUAUUGAAACGCCUAUCGAACUA-UCGAGAGUACGCCGCACUCGCAGUGACGUGUACUC-UUUUUUUUUAGC -------------------.....-.(((((--(((((........))).....)))))))....-..((((((((((..(((.....)))..))))))))-)).......... ( -24.00, z-score = -2.17, R) >droAna3.scaffold_12943 5023413 111 - 5039921 ACAUUAGAGAUGAGCCAUUGCCUU-AGAGAU-GAGCAGUCAAGUCGAAGAGAUGCAAGCACAUCGCUAGCCGAUAGUUCGCGUUUCUAGUGACGUGUACCUUCUUUUUGUUAA- ........(((..((((((.....-...)))-).)).)))....((((((((.....((((.(((((((((((....))).)...))))))).))))....))))))))....- ( -27.70, z-score = -0.64, R) >droEre2.scaffold_4845 19422171 93 - 22589142 --------------CAGCUUCUCC-AUCGAU--GGCCACCAAAUA-AAUCG--CCUAUCGAACUA-UCGCGAGUACGCCGCACUCGCAGUGACGUGUACUCAUUUUUUUUUAGC --------------..(((.....-.(((((--(((.........-....)--)).)))))....-....((((((((..(((.....)))..))))))))..........))) ( -24.72, z-score = -2.66, R) >droYak2.chr2R 2811254 89 - 21139217 ------------------CUCUUC-AUCGAU--AGCCGCAAAGUA-GGAAAACCCUAUCGAACUA-UCGCGAGUACGCCGCACUCGCAGUGACGUGUACUC--UUUUUUUUAGU ------------------......-..((((--((.((....(((-((.....)))))))..)))-))).((((((((..(((.....)))..))))))))--........... ( -25.10, z-score = -2.31, R) >droSec1.super_5 5080509 81 - 5866729 -------------------UCUAC-AUCGAU--AGUCAC---AUC-GAUAG----UAUUGAACUA-UCGCGAGUACGCCGCACUCGCAGUGACGUGUACUC--UUUUUUUUAGC -------------------..(((-((....--.(((((---...-(((((----(.....))))-))((((((.......)))))).))))))))))...--........... ( -23.70, z-score = -2.42, R) >droSim1.chr2L 16064770 81 - 22036055 -------------------UCUAC-AUCGAU--AGUCAC---AUC-GAUAG----UAUCGAACUA-UCGCGAGUACGCCGCACUCGCAGUGACGUGUACUC--UUUUUUUUAGC -------------------.....-..((((--(((...---.((-(((..----.)))))))))-))).((((((((..(((.....)))..))))))))--........... ( -24.80, z-score = -2.67, R) >droWil1.scaffold_180772 4461785 97 - 8906247 --------------CACUCGUUACAAGGGAUGAAACAAUCGAUUAAAGAUGCCAUAGAUUUUUCACGAGCCGCCAC-UAAUUCUUCUUGUGACGUGUACCCUGCCGCAUAUA-- --------------.((.(((((((((((((((((.((((................)))))))))..((......)-)...)).)))))))))).))...............-- ( -16.79, z-score = 0.14, R) >consensus ___________________UCUAC_AUCGAU__AGCCACCAAAUA_AAAAG__CCUAUCGAACUA_UCGCGAGUACGCCGCACUCGCAGUGACGUGUACUC__UUUUUUUUAGC ......................................................................((((((((..(((.....)))..))))))))............. ( -8.47 = -8.80 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:53 2011