
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,339,616 – 16,339,715 |
| Length | 99 |
| Max. P | 0.998914 |

| Location | 16,339,616 – 16,339,713 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.21 |
| Shannon entropy | 0.71858 |
| G+C content | 0.61922 |
| Mean single sequence MFE | -48.36 |
| Consensus MFE | -9.89 |
| Energy contribution | -11.62 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
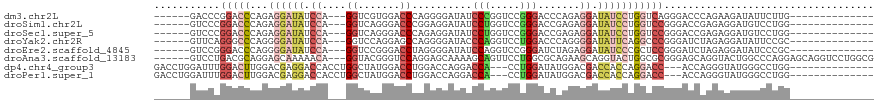
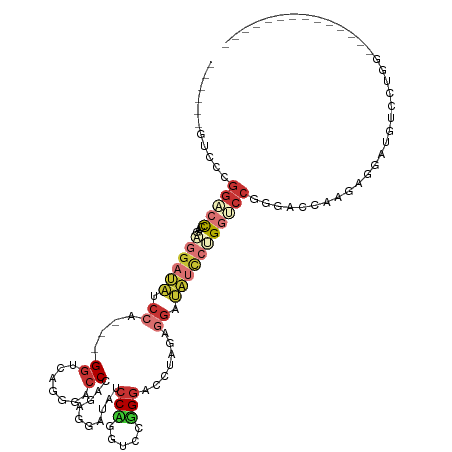
>dm3.chr2L 16339616 97 - 23011544 ------GACCCGGACCCAGAGGAUAUCCA---GGUCGUGGACCCAGGGGAUAUCCCGGUCCGGGACCCAGAGGAUAUCCUGGUCAGGGACCCAGAAGAUAUUCUUG-------------- ------..(((.((((...(((((((((.---((((.((((((...(((....))))))))).))))....))))))))))))).)))....((((....))))..-------------- ( -47.90, z-score = -2.88, R) >droSim1.chr2L 16047289 97 - 22036055 ------GUCCCGGACCCAGAGGAUAUCCA---GGUCAGGGACCCGGAGGAUAUCCUGGUCCGGGACCGAGAGGAUAUCCUGGUCCGGGACCGAGAGGAUGUCCUGG-------------- ------((((((((((...(((((((((.---((((..(((((.(((.....))).)))))..))))....)))))))))))))))))))....(((....)))..-------------- ( -59.10, z-score = -4.30, R) >droSec1.super_5 5063394 97 - 5866729 ------GUCCCGGACCCAGAGGAUAUCCA---GGUCAGGGACCCAGAGGAUAUCCUGGUCCGGGACCGAGAGGAUAUCCUGGUCCGGGACCGAGAGGAUGUCCUGG-------------- ------((((((((((...(((((((((.---((((..(((((...(((....))))))))..))))....)))))))))))))))))))....(((....)))..-------------- ( -56.50, z-score = -4.08, R) >droYak2.chr2R 2794108 97 - 21139217 ------GUUCAGGGCCCAGGGGAUAUCCA---GGUCCAGGAGCCAGGGGAUACCCAGGUCCUGGACCCAGGGGAUAUUCAGGCCCGGGAUCUAGAGGAUAUUCCGC-------------- ------((((.(((((....((((((((.---(((((((((.((.(((....))).)))))))))))....)))))))).))))).)))).....((.....))..-------------- ( -51.10, z-score = -3.58, R) >droEre2.scaffold_4845 19405142 97 - 22589142 ------GUCCGGGACCCAGGGGAUAUCCA---GGUCCGGGACCUAGGGGAUAUCCAGGUCCGGGAUCUAGAGGAUAUCCCGCUCCGGGAUCUAGAGGAUAUCCCGC-------------- ------...((((((((((((((((((((---(((((.((((((.(((....))))))))).))))))...))))))))).))..)))(((.....))).))))).-------------- ( -52.90, z-score = -2.61, R) >droAna3.scaffold_13183 41606 111 + 43642 ------GUCCUGACGCAGGAGCAAAAACA---GGUACGGGUCCAGGAGCAAAAGCAGUUCCUGGCGCAGAAGCAGGUACUGGCGCGGGAGCAGGUACUGGCCCAGGAGCAGGUCCUGGCG ------((((((..((....)).....))---)).))..(.((((((((....)).((((((((.((.......((((((...((....)).)))))).))))))))))...))))))). ( -43.60, z-score = -0.90, R) >dp4.chr4_group3 8039910 100 - 11692001 GACCUGGAUUUGGACUUGGACGAGGACCACCUGGCUAUGGACCUGGACCAGGACCA---CCUGGAUAUGGACGACCACCAGGACC---ACCAGGGUAUGGGCCUGG-------------- ..((.((.......)).)).(.(((.((((((((...(((.(((((.(((((....---)))))...(((....)))))))).))---))))))...))).))).)-------------- ( -37.90, z-score = -1.91, R) >droPer1.super_1 9489342 100 - 10282868 GACCUGGAUUUGGACUUGGACGAGGACCACCUGGCUAUGGACCUGGACCAGGACCA---CCUGGAUAUGGACGACCACCAGGACC---ACCAGGGUAUGGGCCUGG-------------- ..((.((.......)).)).(.(((.((((((((...(((.(((((.(((((....---)))))...(((....)))))))).))---))))))...))).))).)-------------- ( -37.90, z-score = -1.91, R) >consensus ______GUCCCGGACCCAGAGGAUAUCCA___GGUCAGGGACCCAGAGGAUAUCCAGGUCCGGGACCUAGAGGAUAUCCUGGUCCGGGACCAAGAGGAUGUCCUGG______________ ........((.(((((...((((((.((....((.......))..........(((.....))).......)).))))))))))).))................................ ( -9.89 = -11.62 + 1.73)
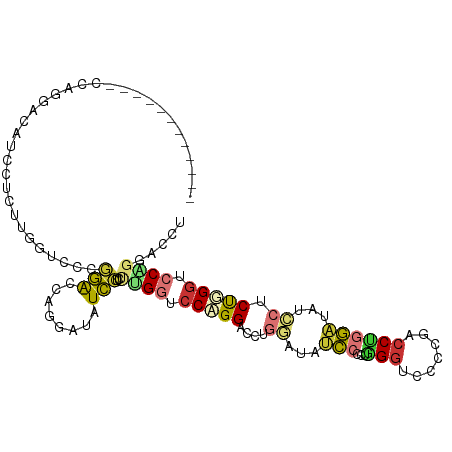
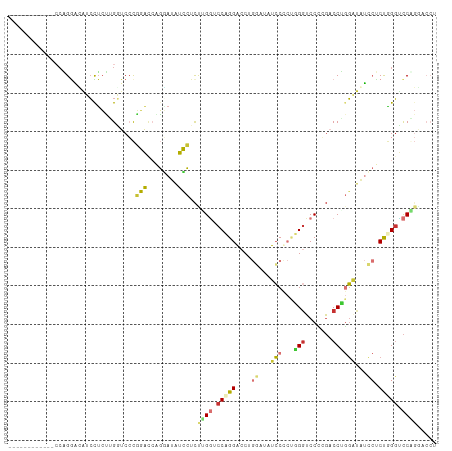
| Location | 16,339,616 – 16,339,715 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 58.27 |
| Shannon entropy | 0.81562 |
| G+C content | 0.61603 |
| Mean single sequence MFE | -47.18 |
| Consensus MFE | -11.85 |
| Energy contribution | -11.54 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
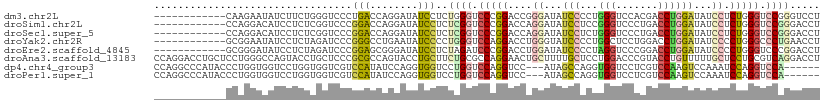
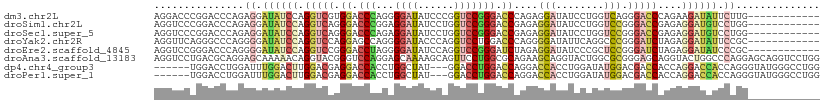
>dm3.chr2L 16339616 99 + 23011544 ------------CAAGAAUAUCUUCUGGGUCCCUGACCAGGAUAUCCUCUGGGUCCCGGACCGGGAUAUCCCCUGGGUCCACGACCUGGAUAUCCUCUGGGUCCGGGUCCU ------------..((((....))))(((.(((.(((((((((((((...(((((..((((((((....)))...)))))..))))))))))))))...)))).))).))) ( -49.40, z-score = -2.38, R) >droSim1.chr2L 16047289 99 + 22036055 ------------CCAGGACAUCCUCUCGGUCCCGGACCAGGAUAUCCUCUCGGUCCCGGACCAGGAUAUCCUCCGGGUCCCUGACCUGGAUAUCCUCUGGGUCCGGGACCU ------------..(((....)))...((((((((((((((((((((....((((..(((((.(((.....))).)))))..)))).)))))))))...))))))))))). ( -59.70, z-score = -4.77, R) >droSec1.super_5 5063394 99 + 5866729 ------------CCAGGACAUCCUCUCGGUCCCGGACCAGGAUAUCCUCUCGGUCCCGGACCAGGAUAUCCUCUGGGUCCCUGACCUGGAUAUCCUCUGGGUCCGGGACCU ------------..(((....)))...((((((((((((((((((((....((((..((((((((....)))...)))))..)))).)))))))))...))))))))))). ( -57.90, z-score = -4.39, R) >droYak2.chr2R 2794108 99 + 21139217 ------------GCGGAAUAUCCUCUAGAUCCCGGGCCUGAAUAUCCCCUGGGUCCAGGACCUGGGUAUCCCCUGGCUCCUGGACCUGGAUAUCCCCUGGGCCCUGAACCU ------------..((.....))..........(((((((.((((((...((((((((((((.(((....))).)).)))))))))))))))).)...))))))....... ( -45.40, z-score = -2.68, R) >droEre2.scaffold_4845 19405142 99 + 22589142 ------------GCGGGAUAUCCUCUAGAUCCCGGAGCGGGAUAUCCUCUAGAUCCCGGACCUGGAUAUCCCCUAGGUCCCGGACCUGGAUAUCCCCUGGGUCCCGGACCU ------------..(((((((((...((.(((.(((...(((.....)))...))).))).)))))))))))..((((((.(((((..(.......)..))))).)))))) ( -49.20, z-score = -2.17, R) >droAna3.scaffold_13183 41608 111 - 43642 CCAGGACCUGCUCCUGGGCCAGUACCUGCUCCCGCGCCAGUACCUGCUUCUGCGCCAGGAACUGCUUUUGCUCCUGGACCCGUACCUGUUUUUGCUCCUGCGUCAGGACCU ((((((.....))))))((..((((..((......))..))))..))...((((((((((...((....))))))))...))))((((....(((....))).)))).... ( -36.20, z-score = -0.80, R) >dp4.chr4_group3 8039910 102 + 11692001 CCAGGCCCAUACCCUGGUGGUCCUGGUGGUCGUCCAUAUCCAGGUGGUCCUGGUCCAGGUCC---AUAGCCAGGUGGUCCUCGUCCAAGUCCAAAUCCAGGUCCA------ (((((.......)))))(((.((((((((.((.((((......)))).).(((.(.(((.((---((......)))).))).).))).).)))...))))).)))------ ( -39.80, z-score = -2.46, R) >droPer1.super_1 9489342 102 + 10282868 CCAGGCCCAUACCCUGGUGGUCCUGGUGGUCGUCCAUAUCCAGGUGGUCCUGGUCCAGGUCC---AUAGCCAGGUGGUCCUCGUCCAAGUCCAAAUCCAGGUCCA------ (((((.......)))))(((.((((((((.((.((((......)))).).(((.(.(((.((---((......)))).))).).))).).)))...))))).)))------ ( -39.80, z-score = -2.46, R) >consensus ____________CCAGGACAUCCUCUUGGUCCCGGACCAGGAUAUCCUCUUGGUCCAGGACCUGGAUAUCCCCUGGGUCCCCGACCUGGAUAUCCUCUGGGUCCAGGACCU .................................(((........)))..((((.(((((....((...(((...(((.......))))))...)).))))).))))..... (-11.85 = -11.54 + -0.31)
| Location | 16,339,616 – 16,339,715 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 111 |
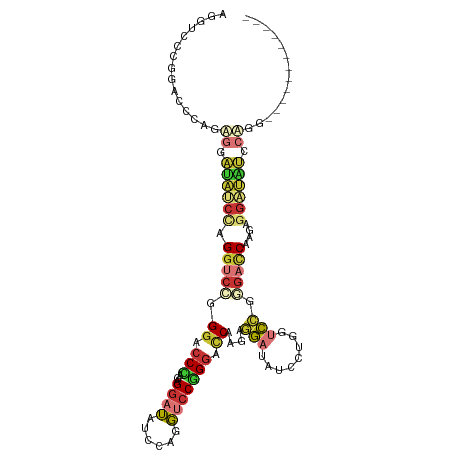
| Reading direction | reverse |
| Mean pairwise identity | 58.27 |
| Shannon entropy | 0.81562 |
| G+C content | 0.61603 |
| Mean single sequence MFE | -50.94 |
| Consensus MFE | -13.45 |
| Energy contribution | -13.71 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.60 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.998914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16339616 99 - 23011544 AGGACCCGGACCCAGAGGAUAUCCAGGUCGUGGACCCAGGGGAUAUCCCGGUCCGGGACCCAGAGGAUAUCCUGGUCAGGGACCCAGAAGAUAUUCUUG------------ .((.(((.((((...(((((((((.((((.((((((...(((....))))))))).))))....))))))))))))).))).)).((((....))))..------------ ( -49.80, z-score = -3.02, R) >droSim1.chr2L 16047289 99 - 22036055 AGGUCCCGGACCCAGAGGAUAUCCAGGUCAGGGACCCGGAGGAUAUCCUGGUCCGGGACCGAGAGGAUAUCCUGGUCCGGGACCGAGAGGAUGUCCUGG------------ .(((((((((((...(((((((((.((((..(((((.(((.....))).)))))..))))....))))))))))))))))))))...(((....)))..------------ ( -63.50, z-score = -5.15, R) >droSec1.super_5 5063394 99 - 5866729 AGGUCCCGGACCCAGAGGAUAUCCAGGUCAGGGACCCAGAGGAUAUCCUGGUCCGGGACCGAGAGGAUAUCCUGGUCCGGGACCGAGAGGAUGUCCUGG------------ .(((((((((((...(((((((((.((((..(((((...(((....))))))))..))))....))))))))))))))))))))...(((....)))..------------ ( -60.90, z-score = -4.89, R) >droYak2.chr2R 2794108 99 - 21139217 AGGUUCAGGGCCCAGGGGAUAUCCAGGUCCAGGAGCCAGGGGAUACCCAGGUCCUGGACCCAGGGGAUAUUCAGGCCCGGGAUCUAGAGGAUAUUCCGC------------ ((((((.(((((....((((((((.(((((((((.((.(((....))).)))))))))))....)))))))).))))).))))))...((.....))..------------ ( -55.30, z-score = -4.51, R) >droEre2.scaffold_4845 19405142 99 - 22589142 AGGUCCGGGACCCAGGGGAUAUCCAGGUCCGGGACCUAGGGGAUAUCCAGGUCCGGGAUCUAGAGGAUAUCCCGCUCCGGGAUCUAGAGGAUAUCCCGC------------ ((((((.(((((..(((....))).))))).))))))..(((((((((((((((.(((....(.((....)))..))).))))))...)))))))))..------------ ( -54.80, z-score = -2.79, R) >droAna3.scaffold_13183 41608 111 + 43642 AGGUCCUGACGCAGGAGCAAAAACAGGUACGGGUCCAGGAGCAAAAGCAGUUCCUGGCGCAGAAGCAGGUACUGGCGCGGGAGCAGGUACUGGCCCAGGAGCAGGUCCUGG .(.(((((...))))).)...........((((.((....((....)).((((((((.((.......((((((...((....)).)))))).)))))))))).)).)))). ( -44.60, z-score = -1.43, R) >dp4.chr4_group3 8039910 102 - 11692001 ------UGGACCUGGAUUUGGACUUGGACGAGGACCACCUGGCUAU---GGACCUGGACCAGGACCACCUGGAUAUGGACGACCACCAGGACCACCAGGGUAUGGGCCUGG ------.((.(((.....(((.(((....)))..)))(((((...(---((.(((((.(((((....)))))...(((....)))))))).))))))))....)))))... ( -39.30, z-score = -1.98, R) >droPer1.super_1 9489342 102 - 10282868 ------UGGACCUGGAUUUGGACUUGGACGAGGACCACCUGGCUAU---GGACCUGGACCAGGACCACCUGGAUAUGGACGACCACCAGGACCACCAGGGUAUGGGCCUGG ------.((.(((.....(((.(((....)))..)))(((((...(---((.(((((.(((((....)))))...(((....)))))))).))))))))....)))))... ( -39.30, z-score = -1.98, R) >consensus AGGUCCCGGACCCAGAGGAUAUCCAGGUCCGGGACCCAGAGGAUAUCCAGGUCCGGGACCAAGAGGAUAUCCUGGUCCGGGACCAAGAGGAUAUCCAGG____________ ...............((.((((((.(((((.((.(((...((((......))))))).))....(((........))).)))))....)))))).)).............. (-13.45 = -13.71 + 0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:48 2011