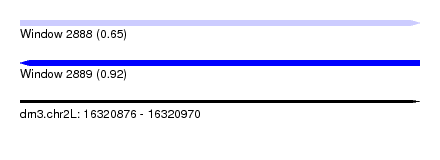
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,320,876 – 16,320,970 |
| Length | 94 |
| Max. P | 0.915394 |

| Location | 16,320,876 – 16,320,970 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 68.87 |
| Shannon entropy | 0.59561 |
| G+C content | 0.31695 |
| Mean single sequence MFE | -16.67 |
| Consensus MFE | -5.30 |
| Energy contribution | -6.13 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
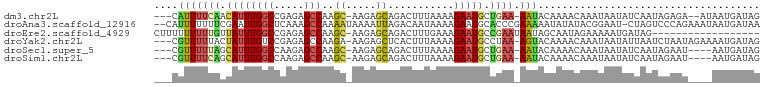
>dm3.chr2L 16320876 94 + 23011544 CUAUCAUUAU--UCUCUAUUGAUAUUAUUUGUUUUGUAUU-UUCAGCAUUCUUUUAAAGUCUGCUCUU-GCUUGGCUCUCGGCCAAAUGUUGAAAAUG--- .(((((.((.--....)).)))))............((((-(((((((((............((....-)).((((.....)))))))))))))))))--- ( -19.90, z-score = -3.39, R) >droAna3.scaffold_12916 9564684 98 - 16180835 UUAUCAUUAUUUCUGGGACUAG-AUUCCGUAUAUAUUUUUCGGGUGCAUUCUUUUAUUGUCUAAUUUUAUUUUGGCUUUGAGCCAAUUCGAAAAAAAUG-- ....((((......((((....-.)))).......(((((((((((((.........)))............((((.....))))))))))))))))))-- ( -15.20, z-score = -0.27, R) >droEre2.scaffold_4929 8015986 82 + 26641161 ------------------CUAUCAUUUUUCUAUUGCUAUUAUUCGGCAUUCUUUCAAAGUCUGCUCUU-GCUUGGCUCUCGGCCAAAUAACAAAAAAAAAG ------------------..............(((.((((....((((..((.....))..))))...-...((((.....)))))))).)))........ ( -10.70, z-score = -0.76, R) >droYak2.chr2L 3317296 96 - 22324452 CUAUCAUUUUCUAUUAGAUUAAUAUUAUUUGUUUUGUACU-UUAGGCAUUCUUUUAAAGUGAGCUCUU-UCUUGGCUCUCGGACAAAUAGUAAAAACG--- ......................((((((((((((...(((-(((((......))))))))(((((...-....)))))..))))))))))))......--- ( -18.50, z-score = -2.32, R) >droSec1.super_5 5044711 92 + 5866729 CUAUCAUU----AUUCUAUUGAUAUUAUUUGUUUUGUAUU-UUCAGCAUUCUUUUAAAGUCUGCUCUU-GCUUGGCUCUUGGCCAAAUGCUAAAAACG--- .(((((..----.......)))))......(((((.....-...((((..((.....))..))))...-(((((((.....)))))..))..))))).--- ( -15.00, z-score = -1.43, R) >droSim1.chr2L 16028324 92 + 22036055 CUAUCAUU----AUUCUAUUGAUAUUAUUUGUUUUGUAUU-UUCAGCAUUCUUUUAAAGUCUGCUCUU-GCUUGGCUCUUGGCCAAAUGCUGAAAACG--- .(((((..----.......)))))..............((-(((((((((............((....-)).((((.....)))))))))))))))..--- ( -20.70, z-score = -3.36, R) >consensus CUAUCAUU____AUUCUAUUAAUAUUAUUUGUUUUGUAUU_UUCAGCAUUCUUUUAAAGUCUGCUCUU_GCUUGGCUCUCGGCCAAAUGCUAAAAACG___ ............................................((((..((.....))..))))......(((((.....)))))............... ( -5.30 = -6.13 + 0.83)


| Location | 16,320,876 – 16,320,970 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 68.87 |
| Shannon entropy | 0.59561 |
| G+C content | 0.31695 |
| Mean single sequence MFE | -17.00 |
| Consensus MFE | -4.55 |
| Energy contribution | -5.25 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16320876 94 - 23011544 ---CAUUUUCAACAUUUGGCCGAGAGCCAAGC-AAGAGCAGACUUUAAAAGAAUGCUGAA-AAUACAAAACAAAUAAUAUCAAUAGAGA--AUAAUGAUAG ---.(((((((.((((((((.....)))..((-....))...........))))).))))-))).............(((((.((....--.)).))))). ( -16.60, z-score = -3.03, R) >droAna3.scaffold_12916 9564684 98 + 16180835 --CAUUUUUUUCGAAUUGGCUCAAAGCCAAAAUAAAAUUAGACAAUAAAAGAAUGCACCCGAAAAAUAUAUACGGAAU-CUAGUCCCAGAAAUAAUGAUAA --((((..((((...(((((.....)))))......((((((........(....)..(((...........)))..)-)))))....)))).)))).... ( -13.00, z-score = -1.16, R) >droEre2.scaffold_4929 8015986 82 - 26641161 CUUUUUUUUUGUUAUUUGGCCGAGAGCCAAGC-AAGAGCAGACUUUGAAAGAAUGCCGAAUAAUAGCAAUAGAAAAAUGAUAG------------------ .((((((.((((((((((((.....)))))..-..(.(((..((.....))..)))).....))))))).)))))).......------------------ ( -15.70, z-score = -1.42, R) >droYak2.chr2L 3317296 96 + 22324452 ---CGUUUUUACUAUUUGUCCGAGAGCCAAGA-AAGAGCUCACUUUAAAAGAAUGCCUAA-AGUACAAAACAAAUAAUAUUAAUCUAAUAGAAAAUGAUAG ---(((((((..(((((((....(((((....-..).))))((((((..........)))-))).....))))))).(((((...)))))))))))).... ( -14.30, z-score = -1.81, R) >droSec1.super_5 5044711 92 - 5866729 ---CGUUUUUAGCAUUUGGCCAAGAGCCAAGC-AAGAGCAGACUUUAAAAGAAUGCUGAA-AAUACAAAACAAAUAAUAUCAAUAGAAU----AAUGAUAG ---.((((((((((((((((.....)))..((-....))...........))))))))))-))).............(((((.......----..))))). ( -20.10, z-score = -3.96, R) >droSim1.chr2L 16028324 92 - 22036055 ---CGUUUUCAGCAUUUGGCCAAGAGCCAAGC-AAGAGCAGACUUUAAAAGAAUGCUGAA-AAUACAAAACAAAUAAUAUCAAUAGAAU----AAUGAUAG ---.((((((((((((((((.....)))..((-....))...........))))))))))-))).............(((((.......----..))))). ( -22.30, z-score = -4.74, R) >consensus ___CGUUUUUACCAUUUGGCCGAGAGCCAAGC_AAGAGCAGACUUUAAAAGAAUGCUGAA_AAUACAAAACAAAUAAUAUCAAUAGAAU____AAUGAUAG ..............((((((.....))))))......(((..((.....))..)))............................................. ( -4.55 = -5.25 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:46 2011