
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 135,480 – 135,598 |
| Length | 118 |
| Max. P | 0.995518 |

| Location | 135,480 – 135,598 |
|---|---|
| Length | 118 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 66.94 |
| Shannon entropy | 0.69505 |
| G+C content | 0.39672 |
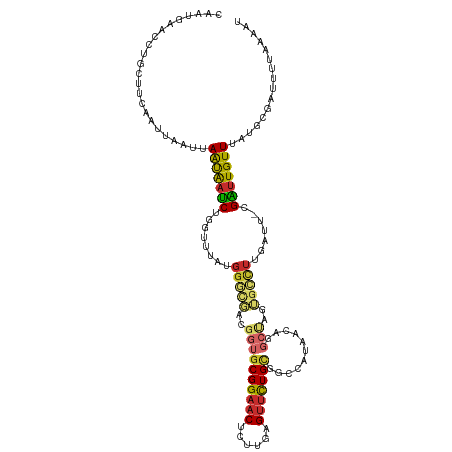
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -15.11 |
| Energy contribution | -14.92 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.995518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
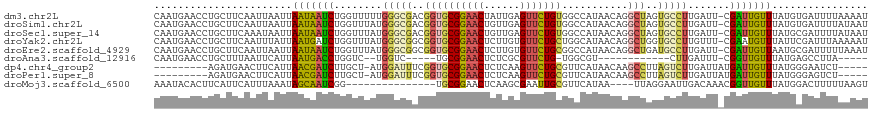
>dm3.chr2L 135480 118 - 23011544 CAAUGAACCUGCUUCAAUUAAUUAAUAAUCUGGUUUUUGGGCGACGGUGCGGAACUAUUGAGUUCUGUGGCCAUAACAGGCUAGUGCCUUGAUU-CGAUUGUUUAUGUGAUUUUAAAAU ...............(((((...(((((((..(((...((((.((...((((((((....))))))))((((......)))).)))))).))).-.)))))))....)))))....... ( -30.20, z-score = -1.67, R) >droSim1.chr2L 138180 118 - 22036055 CAAUGAACCUGCUUCAAUUAAUUAAUAAUCUGGUUUAUGGGCGACGGUGCGGAACUGUUGAGUUCUGUGGCCAUAACAGGCUAGUGCCUUGAUU-CGAUUGUUUAUGUGAUUUUAUAAU ...............(((((...(((((((..(((...((((.((...((((((((....))))))))((((......)))).)))))).))).-.)))))))....)))))....... ( -30.30, z-score = -1.42, R) >droSec1.super_14 134465 118 - 2068291 CAAUGAACCUGCUUCAAAUAAUUAAUAAUCUGGUUUAUGGGCGACGGUGCGGAACUGUUGAGUUCUGUGGCCAUAACAGGCUAGUGCCUUGAUU-CGAUUGUUUAUGCGAUUUUAUAAU ..(((((..(((...........(((((((..(((...((((.((...((((((((....))))))))((((......)))).)))))).))).-.)))))))...)))..)))))... ( -30.54, z-score = -1.30, R) >droYak2.chr2L 124890 118 - 22324452 CAAUGAACCUGCUUCAAUUUAUUAAUGAUCUGGUUUAUGGGCGGCGGUGCGGAACUCUUGUGUUCUGCUGGCAUAACAGGCUGGUGCCUUGUUU-CGAAUGUUUAUUCGAUUUAAAAAU ..(((((((....(((.........)))...))))))).((((.(((((((((((......)))))))((......)).)))).))))(((..(-(((((....))))))..))).... ( -34.20, z-score = -2.40, R) >droEre2.scaffold_4929 170575 118 - 26641161 CAAUGAACCUGCUUCAAUUAAUUAAUAAUCUGGUUUAUGGGCGGCGGUGCGGAACUCUUGUGUUCUGCGGCCAUAACAGGCUGAUGCCUUGAUU-CGAUUGUUAAUGCGAUUUUUAAAU ...((((.....))))....((((((((((..(((...(((((.(...(((((((......)))))))((((......))))).))))).))).-.))))))))))............. ( -34.10, z-score = -2.13, R) >droAna3.scaffold_12916 14207143 93 + 16180835 CAAUGAACCUGCUUUAAUUCAUUAAUGACCUGGUC--UGGUC-----UGCGGAACUCUCGCGUUCUG-UGGCGU------------CUUGAUUU-CGGUUGUUUAUGAGCCUUA----- .................(((((.((..(((.((((--.((.(-----..((((((......))))))-..)...------------)).)))).-.)))..)).))))).....----- ( -23.80, z-score = -1.68, R) >dp4.chr4_group2 160842 104 - 1235136 ---------AGAUGAACUUCAUUAACGAUCUUGCU-AUGGAUUUCGGUGCGGAACUCUCAAGUUCUGCGUUCAUAACAAGCCUUAGUCUUGAUUAUGAUUGUUUAUGGGAAUCU----- ---------((((....(((((.(((((((.....-..(((((..(((((((((((....))))))))((.....))..)))..))))).......))))))).))))).))))----- ( -28.74, z-score = -2.41, R) >droPer1.super_8 237740 104 + 3966273 ---------AGAUGAACUUCAUUAACGAUCUUGCU-AUGGAUUUCGGUGCGGAACUCUCAAGUUCUGCGUUCAUAACAAGCCUUAGUCUUGAUUAUGAUUGUUUAUGGGAGUCU----- ---------....((..(((((.(((((((.....-..(((((..(((((((((((....))))))))((.....))..)))..))))).......))))))).)))))..)).----- ( -29.44, z-score = -2.25, R) >droMoj3.scaffold_6500 258541 100 - 32352404 AAAUACACUUCAUUCAUUUAAAUAGCAAUCGG---------------UGCGGAACUCAAGCGAAUUGCGUUCAUAA----UUAGGAAUUGACAAACGGUUGUUUAUGGACUUUUUAAGU ...............(((((((..(((((...---------------(((.........))).)))))((((((((----...(.(((((.....))))).)))))))))..))))))) ( -16.30, z-score = 0.02, R) >consensus CAAUGAACCUGCUUCAAUUAAUUAAUAAUCUGGUUUAUGGGCGACGGUGCGGAACUCUUGAGUUCUGCGGCCAUAACAGGCUAGUGCCUUGAUU_CGAUUGUUUAUGCGAUUUUAAAAU .......................(((((((........(((((..((((((((((......)))))))...........)))..))))).......)))))))................ (-15.11 = -14.92 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:08 2011