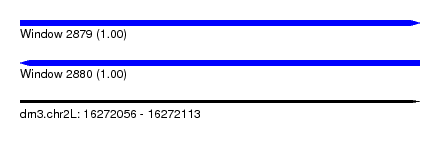
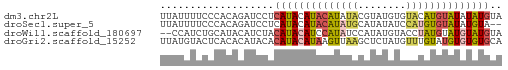
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,272,056 – 16,272,113 |
| Length | 57 |
| Max. P | 0.999015 |

| Location | 16,272,056 – 16,272,113 |
|---|---|
| Length | 57 |
| Sequences | 4 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 62.57 |
| Shannon entropy | 0.61799 |
| G+C content | 0.32153 |
| Mean single sequence MFE | -15.82 |
| Consensus MFE | -9.74 |
| Energy contribution | -11.05 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
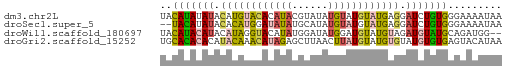
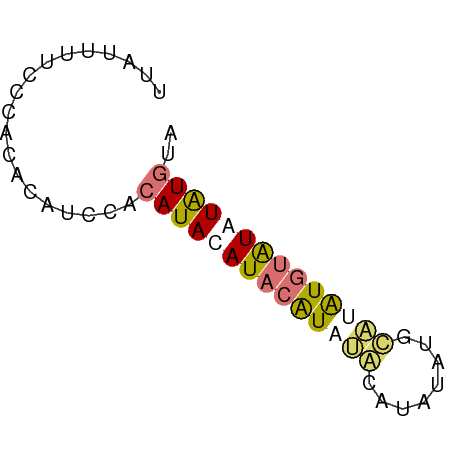
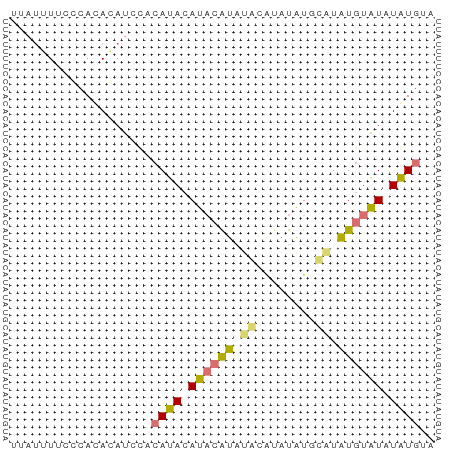
>dm3.chr2L 16272056 57 + 23011544 UACAUAUAUACAUGUACACAUACGUAUAUGUAUGUAUGAGGAUCUGUGGGAAAAUAA ..(((((((((((((((......)))))))))))))))................... ( -18.30, z-score = -3.38, R) >droSec1.super_5 4997603 55 + 5866729 --UACAUAUACACAUGGAUAUAUGCAUAUGUAUGUAUGAGGAUCUGUGGGAAAAUAA --..........(((((((.(((((((....)))))))...)))))))......... ( -11.30, z-score = -1.11, R) >droWil1.scaffold_180697 1807114 55 - 4168966 UACAUACAUACAUAGGUACAUAUGGAUAUGGAUGUAUGUAGAUGUAUGCAGAUGG-- ..(((.(((((((...((((((((........)))))))).)))))))...))).-- ( -14.00, z-score = -1.50, R) >droGri2.scaffold_15252 1331072 57 - 17193109 UGCACACACAUACAAACAUAGAGCUUAACUUAUGUAUGUGUAUGUGUGAGUACAUAA (((.((((((((((.((((((........))))))...)))))))))).)))..... ( -19.70, z-score = -3.05, R) >consensus UACACACAUACACAUACACAUAUGUAUAUGUAUGUAUGAGGAUCUGUGGGAAAAUAA ..(((((((.((((((((((((......)))))))))))).)))))))......... ( -9.74 = -11.05 + 1.31)
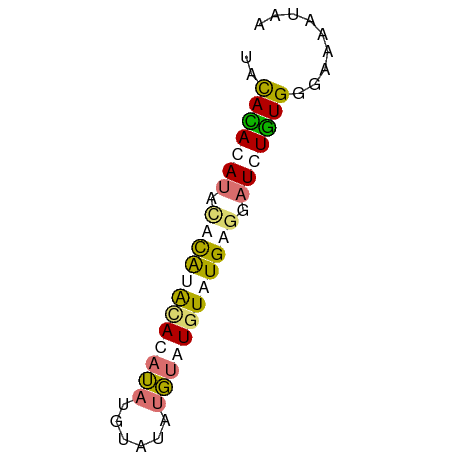
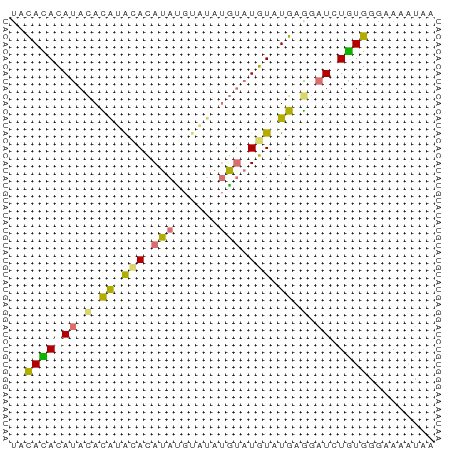
| Location | 16,272,056 – 16,272,113 |
|---|---|
| Length | 57 |
| Sequences | 4 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 62.57 |
| Shannon entropy | 0.61799 |
| G+C content | 0.32153 |
| Mean single sequence MFE | -13.57 |
| Consensus MFE | -8.05 |
| Energy contribution | -8.92 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16272056 57 - 23011544 UUAUUUUCCCACAGAUCCUCAUACAUACAUAUACGUAUGUGUACAUGUAUAUAUGUA .........................((((((((..((((....))))..)))))))) ( -10.60, z-score = -1.24, R) >droSec1.super_5 4997603 55 - 5866729 UUAUUUUCCCACAGAUCCUCAUACAUACAUAUGCAUAUAUCCAUGUGUAUAUGUA-- .........................(((((((((((((....)))))))))))))-- ( -12.70, z-score = -3.31, R) >droWil1.scaffold_180697 1807114 55 + 4168966 --CCAUCUGCAUACAUCUACAUACAUCCAUAUCCAUAUGUACCUAUGUAUGUAUGUA --.....((((((((..((((((....((((....))))....)))))))))))))) ( -12.30, z-score = -2.16, R) >droGri2.scaffold_15252 1331072 57 + 17193109 UUAUGUACUCACACAUACACAUACAUAAGUUAAGCUCUAUGUUUGUAUGUGUGUGCA ...(((((..(((((((((...(((((((.....)).))))).)))))))))))))) ( -18.70, z-score = -2.67, R) >consensus UUAUUUUCCCACACAUCCACAUACAUACAUAUACAUAUAUGCAUAUGUAUAUAUGUA ...................((((((((((((((........)))))))))))))).. ( -8.05 = -8.92 + 0.87)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:39 2011