
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,217,668 – 16,217,762 |
| Length | 94 |
| Max. P | 0.891056 |

| Location | 16,217,668 – 16,217,762 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.94 |
| Shannon entropy | 0.52692 |
| G+C content | 0.41514 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -13.76 |
| Energy contribution | -13.50 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
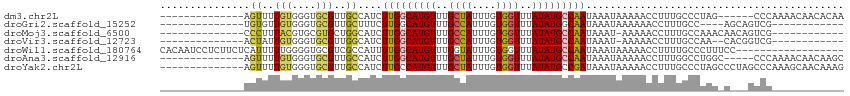
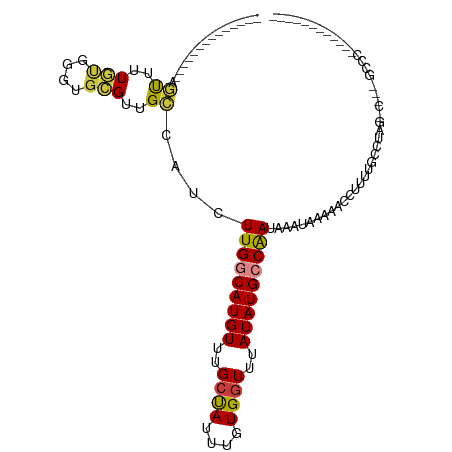
>dm3.chr2L 16217668 94 - 23011544 --------------AGUUUUGUGGGUGCGUUGCCAUCUUGGCAUGUUUGCUAUUUGUGGUUUAUAUGCCAAUAAAUAAAAACCUUUGCCCUAG------CCCAAAACAACACAA --------------.((((((((((.(((.((((.....)))).((((..((((((((((......))).))))))).))))...))))))).------..))))))....... ( -25.70, z-score = -1.43, R) >droGri2.scaffold_15252 8403816 84 + 17193109 --------------UGUGUUGUGGGUGCGUUGCUUUCUUGGCAUGUUUGCCAUUUGUGGUUUAUAUGGCAAUAAAUAAAAAACCUUUGCC----AGCAGUCG------------ --------------..(((((.((((...(((.(((.(((.(((((..((((....))))..))))).))).))))))...))))....)----))))....------------ ( -23.00, z-score = -1.06, R) >droMoj3.scaffold_6500 9722698 87 - 32352404 --------------CCCUUUACGUGCGUGCUGGCAUCUUGGCAUGUUUGCCAUUUGUGGUUUAUAUGCCAAUAAAU-AAAAACCUUUGCCAAACAACAGUCG------------ --------------........((..((..(((((..(((((((((..((((....))))..))))))))).....-.........))))).)).)).....------------ ( -24.19, z-score = -2.13, R) >droVir3.scaffold_12723 4177918 85 + 5802038 --------------ACUAUUGUGGGUGCGUUGGCAUCUUGGCAUGUUUGCCAUUUGUGGUUUAUAUGCCAAUAAAU-AAAAACCUUUGCCAA--CACGGUCG------------ --------------........((.((.(((((((..(((((((((..((((....))))..))))))))).....-.........))))))--).)).)).------------ ( -29.79, z-score = -3.07, R) >droWil1.scaffold_180764 1699312 96 + 3949147 CACAAUCCUCUUCUCAUUUUUGGGGUGCGUCGCCAUUUUGGCAUGUUUGGUAUUUGUGGUUUAUAUGCCAAUAAAUAAAAACCUUUUGCCCUUUCC------------------ .....................(((((.....(((.....)))..((((..((((((((((......))).))))))).)))).....)))))....------------------ ( -20.70, z-score = -0.94, R) >droAna3.scaffold_12916 16003550 95 + 16180835 --------------AGUUUUGUGGGUGCGUUGCCAUCUUGGCAUGUUUGCUAUUUGUGGUUUAUAUGCCAAUAAAUAAAAACCUUUGGCCUGGC-----CCCAAAACAACAAGC --------------.((((((.((((.((..((((..(((((((((..((((....))))..)))))))))..............)))).))))-----))))))))....... ( -30.49, z-score = -1.91, R) >droYak2.chr2L 3208151 100 + 22324452 --------------AGUUUUGUGGGUGCGUUGCCAUCUUGCCAUGUUUGCUAUUUGUGGUUUAUAUGCCGAUAAAUAAAAACCUUUGCCCUAGCCCUAGCCCAAAGCAACAAAG --------------.((((((.((((((((.((......)).))))....((((((((((......))).))))))).........))))..((....)).))))))....... ( -20.40, z-score = 0.32, R) >consensus ______________AGUUUUGUGGGUGCGUUGCCAUCUUGGCAUGUUUGCUAUUUGUGGUUUAUAUGCCAAUAAAUAAAAACCUUUUGCCUAG_C___GCCC____________ ...............((..(((....)))..))....(((((((((..((((....))))..)))))))))........................................... (-13.76 = -13.50 + -0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:32 2011