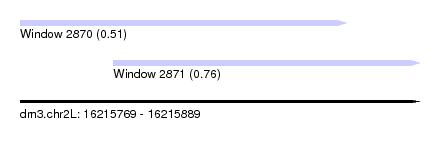
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,215,769 – 16,215,889 |
| Length | 120 |
| Max. P | 0.763640 |

| Location | 16,215,769 – 16,215,867 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 85.33 |
| Shannon entropy | 0.31236 |
| G+C content | 0.39270 |
| Mean single sequence MFE | -21.17 |
| Consensus MFE | -13.74 |
| Energy contribution | -13.71 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.512023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
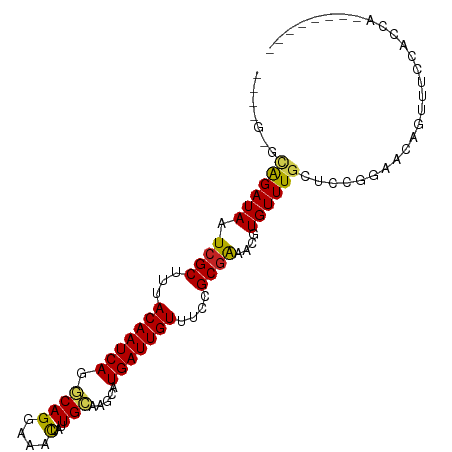
>dm3.chr2L 16215769 98 + 23011544 ---ACAUAUACACGUAUUUAAGCCAUACCUAUAGGGCAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUU ---......((((((......(((..........)))......((((...(((((((.((((....)).......)).)))))))....)))).)))))). ( -26.31, z-score = -2.85, R) >droEre2.scaffold_4929 7912585 98 + 26641161 ---ACACAUGCACAUAUUUAAGCCAUACCUACAGGGCAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUU ---......((((........(((..........)))......((((...(((((((.((((....)).......)).)))))))....))))...)))). ( -23.51, z-score = -1.71, R) >droYak2.chr2L 3206287 91 - 22324452 ----------CACAUAUUUAAGCCAUACAUAUAGAGCAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUU ----------(((........(((.......((((((........))))))......))).((((((.(((........))).)))))).......))).. ( -19.52, z-score = -1.60, R) >droSec1.super_5 4937691 98 + 5866729 ---ACAUAUACACGUAUUUAAGCCAUACCUAUAGGGCAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUU ---......((((((......(((..........)))......((((...(((((((.((((....)).......)).)))))))....)))).)))))). ( -26.31, z-score = -2.85, R) >droSim1.chr2L 15924721 98 + 22036055 ---ACAUAUACACGUAUUUAAGCCAUACCUAUAGGGCAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUU ---......((((((......(((..........)))......((((...(((((((.((((....)).......)).)))))))....)))).)))))). ( -26.31, z-score = -2.85, R) >dp4.chr4_group4 5173702 93 + 6586962 ---GCACAUCCACAUAUUUAAGCCAUACAUA-----UAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGGAACGUGUU ---((((.(((...((((((...........-----))))))...((...(((((((.((((....)).......)).)))))))....)))))..)))). ( -19.01, z-score = -1.12, R) >droPer1.super_16 204152 93 - 1990440 ---GCACAUCCACAUAUUUAAGCCAUACAUA-----UAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGGAACGUGUU ---((((.(((...((((((...........-----))))))...((...(((((((.((((....)).......)).)))))))....)))))..)))). ( -19.01, z-score = -1.12, R) >droAna3.scaffold_12916 16001862 96 - 16180835 ACAGGACACACUCAUUUUUGAGUCAUACAUA-----CAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGGAACGUGUU ....((((((((((....)))))........-----.......((((...(((((((.((((....)).......)).)))))))....))))...))))) ( -24.41, z-score = -2.68, R) >droWil1.scaffold_180764 1697061 94 - 3949147 --GCAAGACUCACAUAUUUAAGCCAUACAUA-----UAGAUAAUCGCUUUACAAUCAGACAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGGAACGUGUU --........(((.((((((...........-----)))))).((((...(((((((...((....))..((....)))))))))....))))...))).. ( -15.90, z-score = -0.35, R) >droGri2.scaffold_15252 8401916 84 - 17193109 -------AUACAUACAUAUAAAUAUUG----------GGAUAAUAGCUUUACAAUCAGACAGGAAGUUAGUGCAAGCAUGAUUGUUUCCGCGAAACGUGUU -------....((((............----------......(((((((............)))))))(((.(((((....))))).))).....)))). ( -11.40, z-score = 0.78, R) >consensus ___ACACAUACACAUAUUUAAGCCAUACAUA_____CAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUU ...........................................((((...(((((((.((((....)).......)).)))))))....))))........ (-13.74 = -13.71 + -0.03)


| Location | 16,215,797 – 16,215,889 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Shannon entropy | 0.37597 |
| G+C content | 0.44108 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -15.37 |
| Energy contribution | -15.08 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16215797 92 + 23011544 --UAGGGCAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUUUGCACCGGAGCAGUUUCCACCA-------- --..(((((((((.((((...(((((((.((((....)).......)).)))))))....))))....))))))).))((((....))))....-------- ( -29.21, z-score = -2.65, R) >droEre2.scaffold_4929 7912613 92 + 26641161 --CAGGGCAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUUUGCACCGGAGCAGUUUCCACCA-------- --..(((((((((.((((...(((((((.((((....)).......)).)))))))....))))....))))))).))((((....))))....-------- ( -29.21, z-score = -2.56, R) >droYak2.chr2L 3206308 92 - 22324452 --UAGAGCAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUUUGCACCGGAGCAGUUUCCACCA-------- --....(((((((.((((...(((((((.((((....)).......)).)))))))....))))....)))))))...((((....))))....-------- ( -26.41, z-score = -2.21, R) >droSec1.super_5 4937719 92 + 5866729 --UAGGGCAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUUUGCACCGGAGCAGUUUCCACCA-------- --..(((((((((.((((...(((((((.((((....)).......)).)))))))....))))....))))))).))((((....))))....-------- ( -29.21, z-score = -2.65, R) >droSim1.chr2L 15924749 92 + 22036055 --UAGGGCAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUUUGCACCGGAGCAGUUUCCACCA-------- --..(((((((((.((((...(((((((.((((....)).......)).)))))))....))))....))))))).))((((....))))....-------- ( -29.21, z-score = -2.65, R) >dp4.chr4_group4 5173730 92 + 6586962 -------UAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGGAACGUGUUUGCUACGGAACGGUUUGAACCGUAGCU--- -------.(((((.((((...(((((((.((((....)).......)).)))))))....))))....)))))(((((((...........))))))).--- ( -26.01, z-score = -1.56, R) >droPer1.super_16 204180 92 - 1990440 -------UAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGGAACGUGUUUGCUGCGGAACGGUUUGAACCGUAGCU--- -------.........(((..(((((((.((((....)).......)).))))))).(((((((..........)))))))(((((....)))))))).--- ( -27.11, z-score = -1.43, R) >droAna3.scaffold_12916 16001893 87 - 16180835 -------CAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGGAACGUGUUUGUUACCGAACAUUUUCAGCCA-------- -------........(((...(((((((.((((....)).......)).)))))))....)))(((.(((((((....))))))).))).....-------- ( -22.41, z-score = -1.84, R) >droWil1.scaffold_180764 1697090 95 - 3949147 -------UAGAUAAUCGCUUUACAAUCAGACAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGGAACGUGUUUGCUGCUUUAACAAUUUUUCCUCCUCCCUU -------.........((.............((....))...(((((((((.....(((....)))))))))))).))........................ ( -15.60, z-score = 0.12, R) >droVir3.scaffold_12723 4175675 94 - 5802038 AUAUGUAGGGAUAAUCGCUUUACAAUCAGACAGGAAGCUAUUGUAAGCAUGAUUGUUUCCGCGAAACGUGUUUGCUACUGCAAAUAUUCUACGA-------- ...((((((.....((((...(((((((.((((.......)))).....)))))))....))))...((((((((....)))))))))))))).-------- ( -26.00, z-score = -2.33, R) >droGri2.scaffold_15252 8401933 89 - 17193109 -----UUGGGAUAAUAGCUUUACAAUCAGACAGGAAGUUAGUGCAAGCAUGAUUGUUUCCGCGAAACGUGUUUGCUGCUGACAACAUUUUGCAA-------- -----...........((.......((......)).(((((((((((((((.((((....))))..))))))))).))))))........))..-------- ( -20.40, z-score = -0.05, R) >consensus ____G_GCAGAUAAUCGCUUUACAAUCAGGCAGGAAACUAUUGCAAGCAUGAUUGUUUCCGCGAAACGUGUUUGCUCCGGAACAGUUUCCACCA________ .......((((((.((((...(((((((.((((....)...))).....)))))))....))))....))))))............................ (-15.37 = -15.08 + -0.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:31 2011