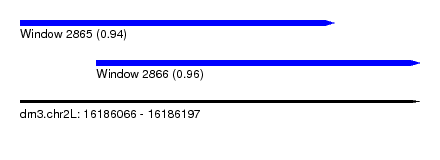
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,186,066 – 16,186,197 |
| Length | 131 |
| Max. P | 0.964122 |

| Location | 16,186,066 – 16,186,169 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.31 |
| Shannon entropy | 0.24820 |
| G+C content | 0.35492 |
| Mean single sequence MFE | -17.15 |
| Consensus MFE | -15.86 |
| Energy contribution | -15.86 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16186066 103 + 23011544 --------AACAUAAACCAAACUUUUACUCAAUCCCCGAA-CAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGC --------................................-.........((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))). ( -16.90, z-score = -1.77, R) >droSim1.chr2L 15894728 103 + 22036055 --------AACAUAAACCAAACUUUUACUCAAUCCCCGAA-CAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGC --------................................-.........((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))). ( -16.90, z-score = -1.77, R) >droSec1.super_5 4907598 103 + 5866729 --------AACAUAAACCAAACUUUUACUCAAUCCCCGAA-CAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGC --------................................-.........((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))). ( -16.90, z-score = -1.77, R) >droYak2.chr2L 3174208 103 - 22324452 --------AACAUAAACCAAACUUUUACUCAAUCCCCGAA-CAAUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGC --------................................-.........((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))). ( -16.90, z-score = -1.82, R) >droEre2.scaffold_4929 7882459 103 + 26641161 --------UACAUAAACCAAACUUUUACUCAAUCCCCGAA-CAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGC --------................................-.........((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))). ( -16.90, z-score = -1.76, R) >droAna3.scaffold_12916 15973669 103 - 16180835 --------AACAUAAACCAAACUUUUACUCAAUCCCCAAA-CAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGC --------................................-.........((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))). ( -16.90, z-score = -2.01, R) >dp4.chr4_group4 5134530 110 + 6586962 -AAAAAAGAACAUAAACCAAACUUUUACUCAAUCCCCGAA-CAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGC -.......................................-.........((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))). ( -16.90, z-score = -1.46, R) >droPer1.super_16 143903 110 - 1990440 -AAAAAAGAACAUAAACCAAACUUUUACUCAAUCCCCGAA-CAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGC -.......................................-.........((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))). ( -16.90, z-score = -1.46, R) >droWil1.scaffold_180764 1650518 87 - 3949147 ----------------------CUUUACUCAAUCCCAAAAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUA---AUGGUAAGC-CUAAAAUGCUGC ----------------------............................((((.(((((....)))))((((((..((..((((...---))))...))-.)))))))))). ( -15.60, z-score = -1.70, R) >droVir3.scaffold_12723 4133615 104 - 5802038 ---------AACAACAACAAAAAUUUACUCAAAUCCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGCGCAAAAAUGCUGC ---------.........................................((((((((((....)))))......((((..((((......)))).....))))...))))). ( -17.60, z-score = -1.42, R) >droMoj3.scaffold_6500 9672133 113 + 32352404 UAAAACAGCAACAACAACAAAAAUUUACUCAAUUCCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGCGCAAAAAUGCUGC .....(((((............................(((.......))).((.(((((....)))))........((..((((......))))...)))).....))))). ( -19.80, z-score = -1.71, R) >droGri2.scaffold_15252 8371244 101 - 17193109 ---------AACAACAACAAA--UUUACUCAAU-CCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAAUAAUGGUAAGCGCAAAAAUGCUGC ---------............--..........-................((((((((((....)))))......((((..((((......)))).....))))...))))). ( -17.60, z-score = -1.37, R) >consensus ________AACAUAAACCAAACUUUUACUCAAUCCCCGAA_CAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC_CUAAAAUGCUGC ..................................................((((.(((((....)))))(((((...((..((((......))))...))...))))))))). (-15.86 = -15.86 + -0.00)

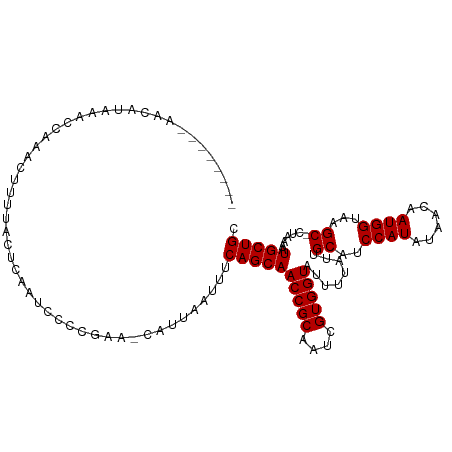
| Location | 16,186,091 – 16,186,197 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 88.53 |
| Shannon entropy | 0.23153 |
| G+C content | 0.33780 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -17.99 |
| Energy contribution | -17.99 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964122 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 16186091 106 + 23011544 CCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAAAAUUAAAACAAAU------- .........(((((((........(((((.(..((((((((..((..((((......))))...))-.))))))))..)..))))).......))))))).......------- ( -20.26, z-score = -2.04, R) >droSim1.chr2L 15894753 106 + 22036055 CCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAAAAUUAAAACAAAU------- .........(((((((........(((((.(..((((((((..((..((((......))))...))-.))))))))..)..))))).......))))))).......------- ( -20.26, z-score = -2.04, R) >droSec1.super_5 4907623 105 + 5866729 CCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAAA-UUAAAACAAAU------- ..........((((((((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))).))))))..........-...........------- ( -20.00, z-score = -1.94, R) >droYak2.chr2L 3174233 113 - 22324452 CCCCGAACAAUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAAAAUUAAAACAUUACAAAAAU ..........((((((((((.(((((....)))))((((((..((..((((......))))...))-.)))))))))).))))))............................. ( -20.10, z-score = -1.95, R) >droEre2.scaffold_4929 7882484 106 + 26641161 CCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAAAAUUAAAACAAAU------- .........(((((((........(((((.(..((((((((..((..((((......))))...))-.))))))))..)..))))).......))))))).......------- ( -20.26, z-score = -2.04, R) >droAna3.scaffold_12916 15973694 103 - 16180835 CCCCAAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAAAAAUUAAAAA---------- .........(((((((........(((((.(..((((((((..((..((((......))))...))-.))))))))..)..)))))........)))))))...---------- ( -20.69, z-score = -2.34, R) >dp4.chr4_group4 5134562 108 + 6586962 CCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAAAAUUAAAUCAAACAA----- .........(((((((........(((((.(..((((((((..((..((((......))))...))-.))))))))..)..))))).......))))))).........----- ( -20.26, z-score = -2.00, R) >droPer1.super_16 143935 108 - 1990440 CCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC-CUAAAAUGCUGCAAAUUGCAAUAAUAAAAUUAAAUCAAACAA----- .........(((((((........(((((.(..((((((((..((..((((......))))...))-.))))))))..)..))))).......))))))).........----- ( -20.26, z-score = -2.00, R) >droVir3.scaffold_12723 4133640 92 - 5802038 CCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGCGCAAAAAUGCUGCAA-UUGCAACAACU--------------------- ........................(((((.(..(((((((..(((..((((......))))...)))..)))))))..).)-)))).......--------------------- ( -21.80, z-score = -2.21, R) >droMoj3.scaffold_6500 9672167 95 + 32352404 CCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGCGCAAAAAUGCUGCAA-UUGCAACAACAACU------------------ ........................(((((.(..(((((((..(((..((((......))))...)))..)))))))..).)-))))..........------------------ ( -21.80, z-score = -2.19, R) >droGri2.scaffold_15252 8371266 92 - 17193109 CCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAAUAAUGGUAAGCGCAAAAAUGCUGCAA-UUGCAACAACU--------------------- ........................(((((.(..(((((((..(((..((((......))))...)))..)))))))..).)-)))).......--------------------- ( -21.80, z-score = -2.23, R) >consensus CCCCGAACAUUAAUUUCAGCAACCGCAAUCGUGGUAUUUUAUUGCAUCCAUAUAACAAUGGUAAGC_CUAAAAUGCUGCAAAUUGCAAUAAUAAAAUUAAAACAAA________ ........................((((..(..(((((((...((..((((......))))...))...)))))))..)...))))............................ (-17.99 = -17.99 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:27 2011