| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,104,462 – 16,104,555 |
| Length | 93 |
| Max. P | 0.997768 |

| Location | 16,104,462 – 16,104,555 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 72.90 |
| Shannon entropy | 0.55347 |
| G+C content | 0.41420 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -17.03 |
| Energy contribution | -16.89 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.997768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
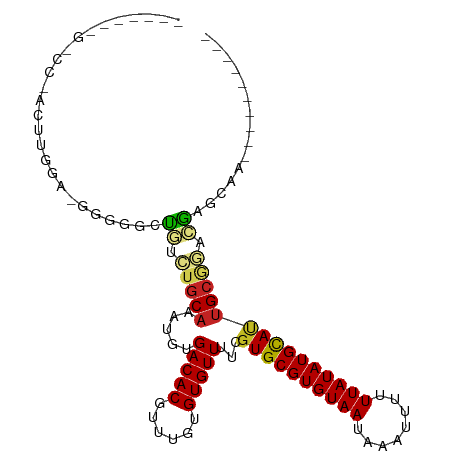
>dm3.chr2L 16104462 93 + 23011544 -----AAGACCUUCUUGGA-GGGGGCUGUCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGGACGAGCAG---------- -----....((((....))-))..((((((((((((((.((((((....))))))..)))((((((((........)))))))).)))))))).)))..---------- ( -33.90, z-score = -3.87, R) >droSim1.chr2L 15824911 93 + 22036055 -----AAGACCGACUUGGA-GGGGGCUGUCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGGACGAGCAG---------- -----....((........-))..((((((((((((((.((((((....))))))..)))((((((((........)))))))).)))))))).)))..---------- ( -31.00, z-score = -2.63, R) >droSec1.super_5 4838147 93 + 5866729 -----AAGACCGACUUGGA-GGGGGCUGUCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGGACGAGCAG---------- -----....((........-))..((((((((((((((.((((((....))))))..)))((((((((........)))))))).)))))))).)))..---------- ( -31.00, z-score = -2.63, R) >droYak2.chr2L 3098691 92 - 22324452 ------AGGCCAACUUGGA-AGGGGCUGUCUGCAAAUGCGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGGACGAGCAG---------- ------...((.....)).-....(((((((((((....((((((....))))))....(((((((((........))))))))))))))))).)))..---------- ( -29.40, z-score = -1.73, R) >droEre2.scaffold_4929 7811071 93 + 26641161 -----ACGACCAACUUGGA-GGGGGCUGUCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGGACGAGCUG---------- -----....((.....)).-...(((((((((((((((.((((((....))))))..)))((((((((........)))))))).)))))))).)))).---------- ( -31.40, z-score = -2.72, R) >droAna3.scaffold_12916 15905901 94 - 16180835 -----AUGGCUGAGCGGAAUGGGGAUUGUCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAGUGCGGACGAACAA---------- -----....................(((((((((.(((.((((((....))))))..)))((((((((........))))))))..)))))))))....---------- ( -26.10, z-score = -1.07, R) >droPer1.super_16 49329 75 - 1990440 --------------------------CUGGGGCAAAUGUGACACGUUUGGGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGUACUGCGCAAGACAAAAA-------- --------------------------((.(.(((.....(((((......)))))...((((((((((........))))))))))))).).)).......-------- ( -19.00, z-score = -1.50, R) >droWil1.scaffold_180764 2388975 99 + 3949147 CUUUCCUUUCUCUAUCAGAAAAAAAGUCCCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGUACAAGCAA---------- ......(((((.....)))))....(((.(.......).)))..((((((((((....((((((((((........)))))))))).))))))))))..---------- ( -23.00, z-score = -2.91, R) >droMoj3.scaffold_6500 9564325 95 + 32352404 --------------AUUGCCUUGCGAUGGUUGCAAAUGCGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUCGAACAAAAAACAGCAACAACAA --------------......((((....(((((....)))))..((((...(((((..((((((((((........)))))))))).)))))..)))).))))...... ( -27.40, z-score = -1.57, R) >droGri2.scaffold_15252 8292013 85 - 17193109 -------------GCCCCCUCCUCGAUGACUGCAAAUGCGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCAAAAAAAAA----------- -------------((........(((.(((((((((((.....)))))))).))).)))(((((((((........)))))))))..)).........----------- ( -20.00, z-score = -2.61, R) >dp4.chr4_group4 5027148 76 + 6586962 --------------------------CUGGGGCAAAUGUGACACGUUUGGGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGUACUGCGCAAGACAAAAAA------- --------------------------((.(.(((.....(((((......)))))...((((((((((........))))))))))))).).))........------- ( -19.00, z-score = -1.53, R) >consensus _______G_CC_ACUUGGA_GGGGGCUGUCUGCAAAUGUGACACGUUUGUGUGUUUUCGUGCGUGUAAUAAAUUUUUUAUAUGCAUUGCGGACGAGCAA__________ ..........................((.(((((.....(((((......)))))...((((((((((........))))))))))))))).))............... (-17.03 = -16.89 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:20 2011