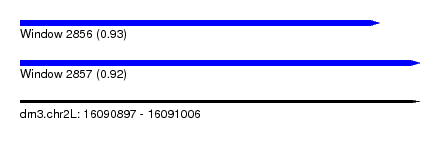
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,090,897 – 16,091,006 |
| Length | 109 |
| Max. P | 0.934480 |

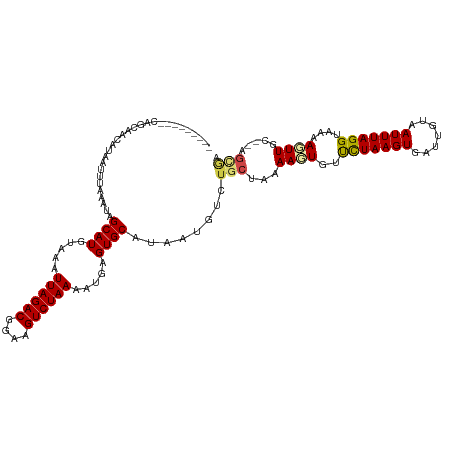
| Location | 16,090,897 – 16,090,995 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.45 |
| Shannon entropy | 0.20973 |
| G+C content | 0.31166 |
| Mean single sequence MFE | -22.61 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.12 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16090897 98 + 23011544 ----------------------CAGGAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAA ----------------------..(((((((..........((((.....((((((....)))))).....)))).....(....).....)))))))...................... ( -19.80, z-score = -1.76, R) >droSim1.chr2L 15809968 98 + 22036055 ----------------------CAGGAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAA ----------------------..(((((((..........((((.....((((((....)))))).....)))).....(....).....)))))))...................... ( -19.80, z-score = -1.76, R) >droSec1.super_5 4824442 98 + 5866729 ----------------------CAGGAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAA ----------------------..(((((((..........((((.....((((((....)))))).....)))).....(....).....)))))))...................... ( -19.80, z-score = -1.76, R) >droYak2.chr2L 3083774 98 - 22324452 ----------------------CAGCAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAA ----------------------.((((((((.((.......((((.....((((((....)))))).....)))))).))).)))))........(((((((.......))))))).... ( -23.51, z-score = -2.76, R) >droEre2.scaffold_4929 7797329 98 + 26641161 ----------------------CAACGGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAA ----------------------....((((....(((....((((.....((((((....)))))).....)))).)))....))))........(((((((.......))))))).... ( -19.50, z-score = -1.60, R) >droAna3.scaffold_12916 15893351 98 - 16180835 ----------------------CAGCAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUCCUAAGUGAUUGUAAUUUAGGUAAA ----------------------.((((((((.((.......((((.....((((((....)))))).....)))))).))).)))))........(((((((.......))))))).... ( -25.71, z-score = -3.64, R) >droVir3.scaffold_12723 4010282 99 - 5802038 --------------------GCUGGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAACUGUUCUAAGUGAUUGUAAUUUAGGAUA- --------------------..(((((((((.((.......((((.....((((((....)))))).....)))))).)))).)))))....((((((((((.......))))))))))- ( -25.01, z-score = -2.87, R) >droMoj3.scaffold_6500 9532261 119 + 32352404 UGUUGGCUUCUUGCUGGAGCACUGGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAACUGUUUUAAGUGAUUGUAAUUUAGGAUA- ..(((((...((((..(....)..)))).............((((.....((((((....)))))).....)))).........)))))...((((((((((.......))))))))))- ( -29.30, z-score = -1.70, R) >droGri2.scaffold_15252 8278305 106 - 17193109 -------------GUUCUGGGCUGGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAGAAUGUCUGCUUAAACUGUUCUAAGUGAUUGUAAUUUAGGAUA- -------------(((.(((((((....))...........((((.....((((((....)))))).....)))).........))))))))((((((((((.......))))))))))- ( -27.70, z-score = -2.65, R) >dp4.chr4_group4 5008356 98 + 6586962 ----------------------CAGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGAAAGUCUAAAACGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAA ----------------------.((((((((.((.......((((.....((((((....)))))).....)))))).)))).))))........(((((((.......))))))).... ( -21.11, z-score = -2.27, R) >droPer1.super_16 30844 98 - 1990440 ----------------------CAGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAACGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAA ----------------------.((((((((.((.......((((.....((((((....)))))).....)))))).)))).))))........(((((((.......))))))).... ( -21.11, z-score = -2.13, R) >droWil1.scaffold_180764 2369774 96 + 3949147 ------------------------CUAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUGGA ------------------------....(((..........((((.....((((((....)))))).....)))).....................((((((.......))))))))).. ( -19.00, z-score = -1.50, R) >consensus ______________________CAGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAA .........................................((((.....((((((....)))))).....))))....................(((((((.......))))))).... (-18.28 = -18.12 + -0.15)

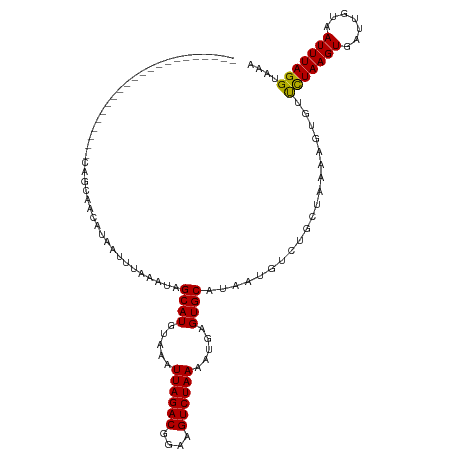
| Location | 16,090,897 – 16,091,006 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.09 |
| Shannon entropy | 0.23014 |
| G+C content | 0.32569 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.26 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16090897 109 + 23011544 ---------CAGGAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAACUUGC--AGCGA ---------...................((((.....((((((....)))))).....))))........((((..((((..(((((((.......)))))))....))))..--)))). ( -26.00, z-score = -2.18, R) >droSim1.chr2L 15809968 109 + 22036055 ---------CAGGAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAACUUGC--AGCGA ---------...................((((.....((((((....)))))).....))))........((((..((((..(((((((.......)))))))....))))..--)))). ( -26.00, z-score = -2.18, R) >droSec1.super_5 4824442 109 + 5866729 ---------CAGGAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAACUUGC--AGCGA ---------...................((((.....((((((....)))))).....))))........((((..((((..(((((((.......)))))))....))))..--)))). ( -26.00, z-score = -2.18, R) >droYak2.chr2L 3083774 102 - 22324452 ---------CAGCAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAACUU--------- ---------.((((((((.((.......((((.....((((((....)))))).....)))))).))).)))))..((((..(((((((.......)))))))....))))--------- ( -25.61, z-score = -3.14, R) >droEre2.scaffold_4929 7797329 109 + 26641161 ---------CAACGGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAACUUGC--AGCGA ---------....(((((.((.......((((.....((((((....)))))).....)))))).)))))((((..((((..(((((((.......)))))))....))))..--)))). ( -27.11, z-score = -2.55, R) >droAna3.scaffold_12916 15893351 109 - 16180835 ---------CAGCAGCAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUCCUAAGUGAUUGUAAUUUAGGUAAAAGUUGC--AGGGA ---------..(((((............((((.....((((((....)))))).....))))....................(((((((.......))))))).....)))))--..... ( -28.10, z-score = -3.04, R) >droVir3.scaffold_12723 4010282 110 - 5802038 -------GCUGGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAACUGUUCUAAGUGAUUGUAAUUUAGG-AUAAGUUGCG-AGCA- -------(((.(((((............((((.....((((((....)))))).....)))).................((((((((((.......)))))))-))).))))).-))).- ( -36.00, z-score = -4.99, R) >droMoj3.scaffold_6500 9532274 112 + 32352404 CUGGAGCACUGGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAACUGUUUUAAGUGAUUGUAAUUUAGG-AUAAGUUGC------- ....((((.((....))...........((((.....((((((....)))))).....))))........))))..(((((((((((((.......)))))))-)).))))..------- ( -26.20, z-score = -1.85, R) >droGri2.scaffold_15252 8278308 114 - 17193109 ---CUGGGCUGGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAGAAUGUCUGCUUAAACUGUUCUAAGUGAUUGUAAUUUAGG-AUAAGUUGCG-CGUA- ---..((((((....))...........((((.....((((((....)))))).....)))).....))))((...(((((((((((((.......)))))))-)).))))..)-)...- ( -30.80, z-score = -2.53, R) >dp4.chr4_group4 5008356 109 + 6586962 ---------CAGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGAAAGUCUAAAACGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAAGUUUC--AGUGA ---------.((((((((.((.......((((.....((((((....)))))).....)))))).)))).))))........(((((((.......)))))))..........--..... ( -21.11, z-score = -1.29, R) >droPer1.super_16 30844 109 - 1990440 ---------CAGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAACGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAAGUUUC--AGUGA ---------.((((((((.((.......((((.....((((((....)))))).....)))))).)))).))))........(((((((.......)))))))..........--..... ( -21.11, z-score = -1.10, R) >droWil1.scaffold_180764 2369774 109 + 3949147 -----------CUAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUGGAAGUUUCUUAGUGA -----------.................((((.....((((((....)))))).....)))).........(((((.((..(((((..(((.......)))..)))))..)).))))).. ( -21.70, z-score = -1.30, R) >consensus _________CAGCAACAUAAUUUAAAUAGCAUGUAAAUUAGACGGAAGUCUAAAAUGAGUGCAUAAUGUCUGCUAAAAGUGUUCUAAGUGAUUGUAAUUUAGGUAAAAGUUGC__AGCGA ............................((((.....((((((....)))))).....))))..............((((..(((((((.......)))))))....))))......... (-19.28 = -19.26 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:19 2011