
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,053,480 – 16,053,544 |
| Length | 64 |
| Max. P | 0.784906 |

| Location | 16,053,480 – 16,053,544 |
|---|---|
| Length | 64 |
| Sequences | 9 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 69.64 |
| Shannon entropy | 0.62048 |
| G+C content | 0.49927 |
| Mean single sequence MFE | -14.76 |
| Consensus MFE | -6.51 |
| Energy contribution | -6.25 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.784906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
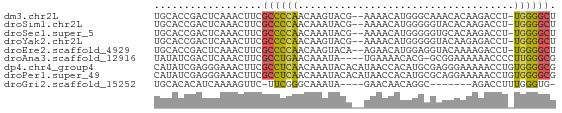
>dm3.chr2L 16053480 64 + 23011544 UGCACCGACUCAAACUUCGCCCCAACAAGUACG--AAAACAUGGGCAAACACAAGACCU-UGGGGCU ..................(((((((((.((...--...)).))(((........).)))-)))))). ( -13.40, z-score = -0.75, R) >droSim1.chr2L 15771211 64 + 22036055 UGCACCGACUCAAACUUCGCCCCAACAAAUACG--AAAACAUGGGGGUACACAAGACCU-UGGGGCU ..................(((((..(......)--........(((((.......))))-)))))). ( -13.00, z-score = -0.06, R) >droSec1.super_5 4786866 64 + 5866729 UGCACCGACUCAAACUUCGCCCCAACAAAUACG--AAAACAUGGGGGUGCACAAGACCU-UGGGGCU .((.((((.........((((((..........--........)))))).........)-))).)). ( -14.44, z-score = -0.12, R) >droYak2.chr2L 3044991 64 - 22324452 UGCACCGACUCAAACUUCGCCCCAACAAGUACG--AAAACAUGGGGGUACAAGAGACCU-UGGGGCU .((.((............(((((..((.((...--...)).))))))).((((....))-)))))). ( -15.70, z-score = -0.33, R) >droEre2.scaffold_4929 7752826 64 + 26641161 UGCACCGACUCAAACUUCGCCCCAACAAGUACA--AGAACAUGGAGGUACAAAAGACCU-UGGGGCU ..................(((((..((.((...--...)).))(((((.......))))-)))))). ( -16.20, z-score = -1.46, R) >droAna3.scaffold_12916 15856886 62 - 16180835 UAUAUCGACUCAAACUUCGCCUGAACAAAUA----UGAAAACACG-GCGGAAAAAACCCCUUGGGCG .................((((..(.((....----)).......(-(.((......)))))..)))) ( -10.20, z-score = -1.02, R) >dp4.chr4_group4 4916841 67 + 6586962 CAUAUCGAGGGAAACUUCGCCUCAACAAAUACACAUAACCACAUGCGAGGGAAAAACCUGUGGGGCG ......((((....))))((((((.......(.(((......))).)(((......))).)))))). ( -20.30, z-score = -3.77, R) >droPer1.super_49 355885 67 - 569410 CAUAUCGAGGGAAACUUCGCCUCAACAAAUACACAUAACCACAUGCGCAGGAAAAACCUGUGGGGCG ......((((....))))(((((..........(((......))).(((((.....)))))))))). ( -21.30, z-score = -4.32, R) >droGri2.scaffold_15252 8956005 54 + 17193109 UGCACACAUCAAAAGUUC-UUCGGGCAAAUA----GAACAACAGGC-------AGACCUUUGGGUG- ....(((.(((((.((((-(..........)----))))....((.-------...))))))))))- ( -8.30, z-score = 0.17, R) >consensus UGCACCGACUCAAACUUCGCCCCAACAAAUACG__AAAACAUGGGCGUACAAAAGACCU_UGGGGCU ..................((((((....................................)))))). ( -6.51 = -6.25 + -0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:15 2011