| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,040,931 – 16,041,021 |
| Length | 90 |
| Max. P | 0.566559 |

| Location | 16,040,931 – 16,041,021 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 71.08 |
| Shannon entropy | 0.60952 |
| G+C content | 0.30119 |
| Mean single sequence MFE | -10.65 |
| Consensus MFE | -6.16 |
| Energy contribution | -6.18 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.23 |
| Mean z-score | -0.47 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
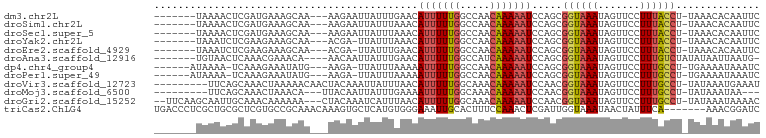
>dm3.chr2L 16040931 90 + 23011544 -------UAAAACUCGAUGAAAGCAA---AAGAAUUAUUUGAACAUUUUUGGCCAACAAAAAUCCAGCGGUAAAUAGUUCCUUUACCU-UAAACACAAUUC -------.................((---(.((((((((((...(((((((.....))))))).......)))))))))).)))....-............ ( -10.50, z-score = -0.36, R) >droSim1.chr2L 15756350 90 + 22036055 -------UAAAACUCGAUGAAAGCAA---AAGAAUUAUUUAAACAUUUUUGGCCAACAAAAAUCCAGCGGUAAAUAGUUCCUUUACCU-UAAACACAAUUC -------.................((---(.((((((((((...(((((((.....))))))).......)))))))))).)))....-............ ( -10.80, z-score = -0.94, R) >droSec1.super_5 4774366 90 + 5866729 -------UAAAACUCGAUGAAAGCAA---AAGAAUUAUUUAAACAUUUUUGGCCAACAAAAAUCCAGCGGUAAAUAGUUCCUUUACCU-UAAACACAAUUC -------.................((---(.((((((((((...(((((((.....))))))).......)))))))))).)))....-............ ( -10.80, z-score = -0.94, R) >droYak2.chr2L 3032431 89 - 22324452 -------UAAAUCUCGAAGAAAGCAA---ACGA-UUAUUUAAACAUUUUUGGCCAACAAAAAUCCAGCGGUAAAUAGUUCCUUUACCU-UAAACACAAUUC -------(((((.(((..........---.)))-..)))))...(((((((.....))))))).....((((((.......)))))).-............ ( -8.10, z-score = 0.02, R) >droEre2.scaffold_4929 7740220 89 + 26641161 -------UAAAUCUCGAAGAAAGCAA---ACGA-UUAUUUGAACAUUUUUGGCCAACAAAAAUCCAGCGGUAAAUAGUUCCUUUACCU-UAAACACAAUUC -------...............((..---.(((-....)))...(((((((.....)))))))...))((((((.......)))))).-............ ( -10.40, z-score = -0.44, R) >droAna3.scaffold_12916 15845156 89 - 16180835 -------UGUAACUCAAACGAAACA----AACAAUUAUUUGAACAUUUUUGGCCAUCAAAAAUCCAGCGGUAAAUAGUUCCUUUGUCUAUAUAAUUAAUG- -------...............(((----((.(((((((((...(((((((.....))))))).......)))))))))..)))))..............- ( -7.70, z-score = 0.71, R) >dp4.chr4_group4 4897411 89 + 6586962 ------AUAAAA-UCAAAGAAAUAUG---AAGA-UUAUUUAAAAAUUUUUGGCCAACAAAAAUCCAGCGGUAAAUAGUUCCUUUGCCU-UGAAAAUAAAUC ------......-(((.(....).))---)...-((((((....(((((((.....))))))).(((.((((((.......)))))))-)).))))))... ( -11.40, z-score = -0.64, R) >droPer1.super_49 336675 89 - 569410 ------AUAAAA-UCAAAGAAAUAUG---AAGA-UUAUUUAAAAAUUUUUGGCCAACAAAAAUCCAGCGGUAAAUAGUUCCUUUGCCU-UGAAAAUAAAUC ------......-(((.(....).))---)...-((((((....(((((((.....))))))).(((.((((((.......)))))))-)).))))))... ( -11.40, z-score = -0.64, R) >droVir3.scaffold_12723 3919414 91 - 5802038 ---------UUCAGCAAACUAAAAACAACUACAAAUUAUUUAACAUUUUUGGCAAACAAAAAUCCAACGGUAAAUAGUUCCUUUGCCU-UAUAAAUGAAAU ---------..........................(((((((..(((((((.....))))))).....((((((.......)))))).-..)))))))... ( -11.10, z-score = -1.60, R) >droMoj3.scaffold_6500 9447324 85 + 32352404 ---------UUCAGCAAACUAAACA---UUACAAUUAUUUGAAAAUUUUUGGCAAACAAAAAUCCAACGGUAAAUAGUUCCUUUGCCU-UAUAAAUAA--- ---------................---......(((((((...(((((((.....))))))).....((((((.......)))))).-..)))))))--- ( -9.90, z-score = -0.83, R) >droGri2.scaffold_15252 8219574 95 - 17193109 --UUCAAGCAAUUGCAAACAAAAAA---CUACAAAUCAUUUAACAUUUUUGGCAAACAAAAAUCCAACGGUAAAUAGUUCCUUUGCCU-UAUAAAUAAAAC --.....((....))..........---.........(((((..(((((((.....))))))).....((((((.......)))))).-..)))))..... ( -10.20, z-score = -1.00, R) >triCas2.ChLG4 6646528 94 - 13894384 UGACCCUCGCUGCGCUCGUGCCGCAAACAAAGUGCUCAUGUGGGAAAUUGCACUUUCCAAACUCGAUUGGUAAAUAACUAUUUCA-------AAACGGAUC ....((..((.(((....))).))....(((((((..............)))))))((((......))))...............-------....))... ( -15.44, z-score = 1.09, R) >consensus _______UAAAACUCAAAGAAAGCAA___AAGAAUUAUUUAAACAUUUUUGGCCAACAAAAAUCCAGCGGUAAAUAGUUCCUUUACCU_UAAAAACAAUUC ............................................(((((((.....))))))).....((((((.......)))))).............. ( -6.16 = -6.18 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:13 2011