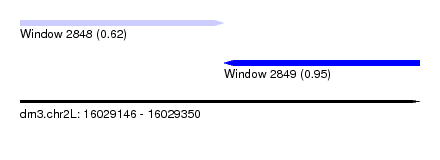
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,029,146 – 16,029,350 |
| Length | 204 |
| Max. P | 0.951832 |

| Location | 16,029,146 – 16,029,250 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 71.61 |
| Shannon entropy | 0.53817 |
| G+C content | 0.38759 |
| Mean single sequence MFE | -14.16 |
| Consensus MFE | -8.78 |
| Energy contribution | -8.62 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.619862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
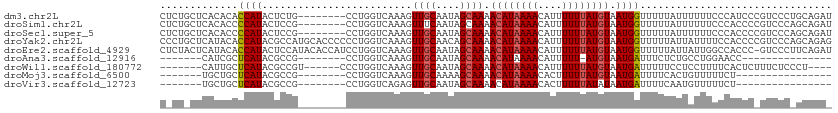
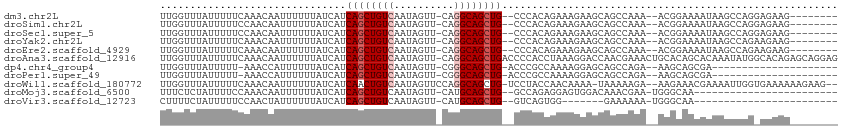
>dm3.chr2L 16029146 104 + 23011544 CUCUGCUCACACACCAUACUCUG--------CCUGGUCAAAGUUGCAAUAGCAAAACAUAAAACAUUUUUUAUGUAAUGGUUUUUAUUUUUUCCCAUCCCGUCCCUGCAGAU ...................((((--------(..((.(....((((....)))).((((((((....)))))))).((((.............))))...).))..))))). ( -17.72, z-score = -1.56, R) >droSim1.chr2L 15744482 104 + 22036055 CUCUGCUCACACCCCAUACUCCG--------CCUGGUCAAAGUUUCAAUAGCAAAACAUAAAACAUUUUUUAUGUAAUGGUUUUUAUUUUUUCCCACCCCGUCCCAGCAGAU .((((((.((............(--------(.(((........)))...))...((((((((....)))))))).........................))...)))))). ( -13.40, z-score = -0.90, R) >droSec1.super_5 4762498 104 + 5866729 CUCUGCUCACACCCCAUACUCCG--------CCUGGUCAAAGUUGCAAUAGCAAAACAUAAAACAUUUUUUAUGUAAUGGUUUUUAUUUUUUCCCACCCCGUCCCAGCAGAU .((((((.((...(((.......--------..)))(((...((((....)))).((((((((....))))))))..)))....................))...)))))). ( -15.20, z-score = -0.98, R) >droYak2.chr2L 3020567 112 - 22324452 CCCUGCUCAUACACCAUACGCCAUGCACCCCCCUGGUCAAAGUUGCAACAGCAAAACAUAAAACAUUUUUUAUGUAAUGGUUUUUAUUAUUUUCCACCCCGUCCCAGCAGAG ..(((((...(((((((..((((.(......).)))).....((((....)))).((((((((....)))))))).)))))...................))...))))).. ( -17.30, z-score = -1.05, R) >droEre2.scaffold_4929 7728519 111 + 26641161 CUCUACUCAUACACCAUACUCCAUACACCAUCCUGGUCAAAGUUGCAAUAGCAAAACAUAAAACAUUUUUUAUGUAAUGGUUUUUAUUAUUGGCCACCC-GUCCCUUCAGAU .................................(((((((..((((....)))).((((((((....))))))))..............)))))))...-............ ( -15.80, z-score = -0.96, R) >droAna3.scaffold_12916 15833760 81 - 16180835 -------CAUCGCUCAUACGCCG--------CCUGGUCAAAGUUGCAAUAGCAAAACAUAAAACAUUUUU-AUGUAAUGAUUUCUCUGCCUGGAACC--------------- -------....((......))..--------((.((.((.((((((....)))).(((((((.....)))-))))........)).)))).))....--------------- ( -12.70, z-score = -0.05, R) >droWil1.scaffold_180772 6565381 95 + 8906247 -------CAUUGCUCAUACGCCGU------CCCUGGUCAAAGUUGCAAUAGCAAAACAUAAAACAUUUUUUAUGUAAUGAUUUUUCCUCCUUUUCACUCUUUCUCCCU---- -------.(((((......((((.------...)))).......)))))..((..((((((((....))))))))..)).............................---- ( -11.22, z-score = -1.31, R) >droMoj3.scaffold_6500 9429512 81 + 32352404 -------UGCUGCUCAUACGCCG--------CCUGGUCAAAGUUGCAAAAGCAAAACAUAAAACACUUUUUAUGUAAUGAUUUUCACUGUUUUUCU---------------- -------.((.((......)).)--------)..(((((...((((....)))).((((((((....))))))))..)))))..............---------------- ( -12.70, z-score = -0.05, R) >droVir3.scaffold_12723 3903179 81 - 5802038 -------UGCUGCUCAUACGCCG--------CCUGGUCAGAGUUGCAAUAGCAAAACAUAAAACACUUUUUAUAUAAUGAUUUUCAAUGUUUUUCU---------------- -------(((.((((....(((.--------...)))..)))).)))..((.(((((((((((....))))))....((.....))..))))).))---------------- ( -11.40, z-score = 0.33, R) >consensus C_CU_CUCACACCCCAUACGCCG________CCUGGUCAAAGUUGCAAUAGCAAAACAUAAAACAUUUUUUAUGUAAUGGUUUUUAUUUUUUUCCACCC_GUCCC_GCAGA_ .............((((.........................((((....)))).((((((((....)))))))).))))................................ ( -8.78 = -8.62 + -0.16)
| Location | 16,029,250 – 16,029,350 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.47 |
| Shannon entropy | 0.55285 |
| G+C content | 0.39102 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -9.66 |
| Energy contribution | -9.94 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
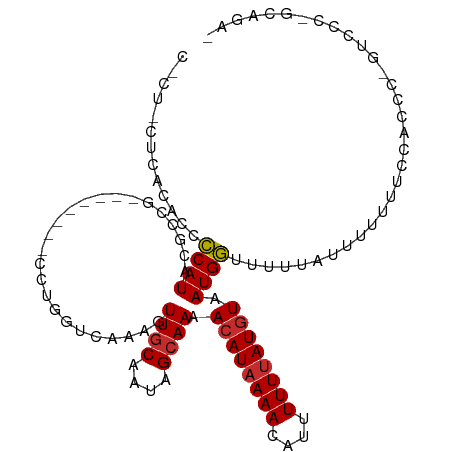
>dm3.chr2L 16029250 100 - 23011544 UUGGUUUAUUUUUCAAACAAUUUUUUAUCAUCAGCUGUCAAUAGUU-CAGGCAGCUG--CCCACAGAAAGAAGCAGCCAAA--ACGGAAAAUAAGCCAGGAGAAG-------- ((((((((((((((......((((((.....((((((((.......-..))))))))--.......))))))...(.....--.)))))))))))))))......-------- ( -26.00, z-score = -3.21, R) >droSim1.chr2L 15744586 100 - 22036055 UUGGUUUAUUUUUCCAACAAUUUUUUAUCAUCAGCUGUCAAUAGUU-CAGGCAGCUG--CCCACAGAAAGAAGCAGCCAAA--ACGGAAAAUAAGCCAGGAGAAG-------- ((((((((((((((......((((((.....((((((((.......-..))))))))--.......)))))).........--..))))))))))))))......-------- ( -25.23, z-score = -2.76, R) >droSec1.super_5 4762602 100 - 5866729 UUGGUUUAUUUUUCCAACAAUUUUUUAUCAUCAGCUGUCAAUAGUU-CAGGCAGCUG--CCCACAGAAAGAAGCAGCCAAA--ACGGAAAAUAAGCCAGGAGAAG-------- ((((((((((((((......((((((.....((((((((.......-..))))))))--.......)))))).........--..))))))))))))))......-------- ( -25.23, z-score = -2.76, R) >droYak2.chr2L 3020679 100 + 22324452 UUGGUUUAUUUUUCAAACAAUUUUUUAUCAUCAGCUGUCAAUAGUU-CAGGCAGCUG--CCCACAGAAAGAAGCAGCCAAA--ACGGAAAAUAAGCCAGAAGAAG-------- ((((((((((((((......((((((.....((((((((.......-..))))))))--.......))))))...(.....--.)))))))))))))))......-------- ( -26.00, z-score = -3.62, R) >droEre2.scaffold_4929 7728630 100 - 26641161 UUGGUUUAUUUUUCAAACAAUUUUUUAUCAUCAGCUGUCAAUAGUU-CAGGCAGCUG--CCCACAGAAAGAAGCAGCCAAA--ACGGAAAAUAAGCCAGAAGAAG-------- ((((((((((((((......((((((.....((((((((.......-..))))))))--.......))))))...(.....--.)))))))))))))))......-------- ( -26.00, z-score = -3.62, R) >droAna3.scaffold_12916 15833841 112 + 16180835 UUGGUUUAUUUUUCAAACAAUUUUUUAUCAUCAGCUGUCAAUAGUU-CAGGCAGCUGACCCCACCUAAAGGACCAACGAAACUGCACAGCACAAAUAUGGCACAGAGCAGGAG (((((.........................(((((((((.......-..))))))))).....((....))))))).....((((.(.((.((....))))...).))))... ( -24.50, z-score = -1.37, R) >dp4.chr4_group4 4883091 87 - 6586962 UUGGUUUAUUUUU-AAACCAUUUUUUAUCAUCAGCUGUCAAUAGUU-CGGGCAGCUG-ACCCGCCAAAAGGAGCAGCCAGA--AAGCAGCGA--------------------- .(((((((....)-))))))..........(((((((((.......-..))))))))-).((.......)).((.((....--..)).))..--------------------- ( -24.20, z-score = -2.14, R) >droPer1.super_49 321840 87 + 569410 UUGGUUUAUUUUU-AAACCAUUUUUUAUCAUCAGCUGUCAAUAGUU-CGGGCAGCUG-ACCCGCCAAAAGGAGCAGCCAGA--AAGCAGCGA--------------------- .(((((((....)-))))))..........(((((((((.......-..))))))))-).((.......)).((.((....--..)).))..--------------------- ( -24.20, z-score = -2.14, R) >droWil1.scaffold_180772 6565476 107 - 8906247 UUGGUUUAUUUUUCAAACAAUUUUUUAUCAUCAACUGUCAAUAGUUCCAGGCAGCUG-UCCUACCAACAAAA-UAAAAAGA--AAGAAACGAAAAUUGGUGAAAAAAGAAG-- ....(((.(((((((..((((((((..((.....(((((..........))))).((-(.......)))...-........--..))...)))))))).))))))).))).-- ( -13.80, z-score = 0.34, R) >droMoj3.scaffold_6500 9429593 85 - 32352404 UUUCUCUAUUUUCCAAACAAUUUUUUAUCAUCAGCUGUCAAUAGUU-CAUGCAGCUG--GCCAGAGGAGUGGACAAACGAA-UGGGCAA------------------------ ....(((((((((((.....((((((....(((((((.((......-..))))))))--)..)))))).)))).....)))-))))...------------------------ ( -17.20, z-score = -0.16, R) >droVir3.scaffold_12723 3903260 78 + 5802038 CUUUUCUAUUUUUCCAACUAUUUUUUAUCAUCAGCUGUCAAUAGUU-CAUGCAGCUG--GUCAGUGG-------GAAAAAA-UGGGCAA------------------------ ....(((((((((((.(((..........((((((((.((......-..))))))))--)).))).)-------).)))))-))))...------------------------ ( -18.70, z-score = -2.46, R) >consensus UUGGUUUAUUUUUCAAACAAUUUUUUAUCAUCAGCUGUCAAUAGUU_CAGGCAGCUG__CCCACAGAAAGAAGCAGCCAAA__ACGCAAAAUAAGCCAG_AGAAG________ ...............................((((((((..........))))))))........................................................ ( -9.66 = -9.94 + 0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:12 2011