
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,989,181 – 15,989,271 |
| Length | 90 |
| Max. P | 0.583306 |

| Location | 15,989,181 – 15,989,271 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 64.37 |
| Shannon entropy | 0.69137 |
| G+C content | 0.47489 |
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -6.12 |
| Energy contribution | -5.60 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.583306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
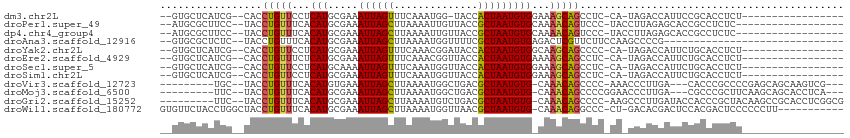
>dm3.chr2L 15989181 90 - 23011544 --GUGCUCAUCG--CACCUGUUCCUCAUGCGAAAUUAGUUUCAAAUGG-UACCACUAAUGUGGAAAGCAGCCUC-CA-UAGACCAUUCCGCACCUCU----------------- --((((...(((--((..((.....)))))))...........(((((-(........((((((........))-))-)).))))))..))))....----------------- ( -20.50, z-score = -1.95, R) >droPer1.super_49 268417 91 + 569410 --AUGCGCUUCC--UACCUGUUUCACAUGCGAAAUUAGCUUAAAAUUGUUACCGCUAAUGUGCAAAACAGUCCC-UACCUUAGAGCACCGCCUCUC------------------ --.(((.((...--...((((((((((((((....((((........)))).)))..)))))..))))))....-......)).))).........------------------ ( -14.69, z-score = -0.95, R) >dp4.chr4_group4 4831102 91 - 6586962 --AUGCGCUUCC--UACCUGUUUCACAUGCGAAAUUAGCUUAAAAUUGUUACCGCUAAUGUGCAAAACAGUCCC-UACCUUAGAGCACCGCCUCUC------------------ --.(((.((...--...((((((((((((((....((((........)))).)))..)))))..))))))....-......)).))).........------------------ ( -14.69, z-score = -0.95, R) >droAna3.scaffold_12916 15797931 80 + 16180835 --GUGCGCUCUC--UACCUGUUUCACAUGCGAAAUUAGCUUAAAAUGGUUUUCGCUAAUGUGAGACUCGUUCUUCCAAGCCCCG------------------------------ --..((......--.((..(((((((((((((((...(((......)))))))))..)))))))))..))........))....------------------------------ ( -19.96, z-score = -2.81, R) >droYak2.chr2L 2979768 91 + 22324452 --GUGCUCAUCG--CACCUGUUCCUCAUGCGAAAUUAGUUUCAAACGGAUACCACUAAUGUGGCAAGCAGCCCC-CA-UAGACCAUUCUGCACCUCU----------------- --((((.....)--)))((((......(((((((....)))).........((((....))))...))).....-.)-)))................----------------- ( -16.10, z-score = -0.10, R) >droEre2.scaffold_4929 7686376 91 - 26641161 --GUGCUCAUCG--CACCUGUUUCUCAUGCGAAAUUAGUUUCAAACGGUUACCACUAAUGUGAAAAGCAGCCUC-CA-UAGACCAUUCUGCACCUCU----------------- --((((...(((--((..((.....)))))))..............((((........(((.....))).....-..-..)))).....))))....----------------- ( -14.93, z-score = -0.29, R) >droSec1.super_5 4722601 91 - 5866729 --GUGCUCAUCG--CACCUGUUCCUCAUGCAAAAUUAGUUUCAAAUGGUUACCACUAAUGUGGAAAGCAGCCUC-CA-UAGACCAUUCUGCACCUCU----------------- --((((.....(--((..((.....))))).............(((((((........((((((........))-))-)))))))))..))))....----------------- ( -17.00, z-score = -0.66, R) >droSim1.chr2L 15703718 91 - 22036055 --GUGCUCAUCG--CACCUGUUCCUCAUGCGAAAUUAGUUUCAAAUGGUUACCACUAAUGUGGAAAGCAGCCUC-CA-UAGACCAUUCUGCACCUCU----------------- --((((...(((--((..((.....)))))))...........(((((((........((((((........))-))-)))))))))..))))....----------------- ( -20.40, z-score = -1.63, R) >droVir3.scaffold_12723 3838238 95 + 5802038 ---------UGC--UACCUGUUUCACAUGUGAAAUUAGCUUAAAAUGGCUGACGCUAAUGUG-CAAACAGCCCC-AAACCCUUGA---CACCCGCCCCGAGCAGCAAGUCG--- ---------(((--(..((((((((((((((...((((((......)))))))))..)))))-.))))))....-..........---.....((.....)))))).....--- ( -21.80, z-score = -1.72, R) >droMoj3.scaffold_6500 9368556 96 - 32352404 ---------UUC--UACCUGUUUCACAUGCGAAAUUAGCUUAAAAUGGCUGACGCUAAUGUG-CAAACAGCCCCGGAACCCUUGA---CGCCCGCUUCAAGCAGCACCUCA--- ---------...--...((((((((((((((...((((((......)))))))))..)))))-.))))))....((....(((((---.(....).))))).....))...--- ( -24.20, z-score = -1.92, R) >droGri2.scaffold_15252 8160316 101 + 17193109 ---------UUC--UACCUGUUUCACAUGCGAAAUUAGCUUAAAAUGUCUGACGCUAAUGUG-CAAACAGCCCC-AAGCCCUUGAUACCACCCGCUACAAGCCGCACCUCGGCG ---------...--...((((((((((((((...(((((.......).)))))))..)))))-.))))))....-.(((...((....))...)))....((((.....)))). ( -21.10, z-score = -1.65, R) >droWil1.scaffold_180772 6504020 100 - 8906247 GUGUUCUACCUGGCUACCUGUUUCACAUGCGAAAUUAGCUUAAAAUGGUUAACGCUAAUGUG-CAAACAGGCCC-CU-GACACGACUCCACGACUCCCCCCUU----------- (((((......((...(((((((((((((((...((((((......)))))))))..)))))-.)))))))..)-).-)))))....................----------- ( -25.00, z-score = -2.91, R) >consensus __GUGCUCAUCC__UACCUGUUUCACAUGCGAAAUUAGCUUAAAAUGGUUACCGCUAAUGUG_AAAACAGCCCC_CA_CACACCAUUCCGCACCUCU_________________ .................(((((...(((.....((((((..............)))))))))...)))))............................................ ( -6.12 = -5.60 + -0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:05 2011