
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,986,517 – 15,986,608 |
| Length | 91 |
| Max. P | 0.532882 |

| Location | 15,986,517 – 15,986,608 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.85 |
| Shannon entropy | 0.46028 |
| G+C content | 0.48158 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -17.06 |
| Energy contribution | -16.81 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.532882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15986517 91 - 23011544 -----GUCAGCCACUCGUCGGCACACUCAUUAUUGGGCUCAAGUGCAAAUUAGUGUGUUUGCUAGUGGGUUAUUAAGGCCUCCCCUGGAAACUCAG------------- -----.....(((((.((..(((((((.(((..((.((....)).))))).)))))))..)).))))).......(((.....)))(....)....------------- ( -25.30, z-score = -0.75, R) >droSim1.chr2L 15701032 91 - 22036055 -----GUCAGCCACUCGUCGGCACACUCAUUAUUGGGCUCAAGUGCAAAUUAGUGUGUUUGCUAGUGGGUUAUUAAGGCCUCCCCUGGAAACUCAG------------- -----.....(((((.((..(((((((.(((..((.((....)).))))).)))))))..)).))))).......(((.....)))(....)....------------- ( -25.30, z-score = -0.75, R) >droEre2.scaffold_4929 7683773 91 - 26641161 -----GUCAGCCACUCGUCGGCACACUCAUUAUUGGGCUCAAGUGCAAAUUAGUGUGUUUGCUAGUGGGUUAUUAAGGCCUCCCCUGGAAACUCAG------------- -----.....(((((.((..(((((((.(((..((.((....)).))))).)))))))..)).))))).......(((.....)))(....)....------------- ( -25.30, z-score = -0.75, R) >droYak2.chr2L 2977104 91 + 22324452 -----GUCAGCCACUCGUCGGCACACUCAUUAUUGGGCUCAAGUGCAAAUUAGUGUGUUUGCUAGUGGGUUAUUAAGGCCUCCCCUGGAAACUCAG------------- -----.....(((((.((..(((((((.(((..((.((....)).))))).)))))))..)).))))).......(((.....)))(....)....------------- ( -25.30, z-score = -0.75, R) >droAna3.scaffold_12916 15795460 104 + 16180835 -----GCCAUCCACUCGUCGGCACACUCAUUAUUGGGCUCAAGUGCAAAUUAGUGUGUUUGCUAGUGGGUUAUUAAGACCCUCCCCCGACGACCACCCGGCCACUCCGC -----(((......(((((((((((((.(((..((.((....)).))))).)))))).......(.(((((.....))))).)..)))))))......)))........ ( -31.30, z-score = -1.68, R) >dp4.chr4_group4 4827583 92 - 6586962 -----CUCGGGCACUCGUCAGCACACUCAUUAUUAGGCUCAAGUGCAAAUUAGUGUGUUUGGUAGUGGGUUAUUAAGUUCUUGCCCCGCUCCCCAAC------------ -----...(((((...........((((((((((((((.((.((....))...)).))))))))))))))...........)))))...........------------ ( -22.75, z-score = -0.52, R) >droPer1.super_49 264883 92 + 569410 -----CUCGGGCACUCGUCAGCACACUCAUUAUUAGGCUCAAGUGCAAAUUAGUGUGUUUGGUAGUGGGUUAUUAAGUUCUUGCCCCGCUCCCCAAC------------ -----...(((((...........((((((((((((((.((.((....))...)).))))))))))))))...........)))))...........------------ ( -22.75, z-score = -0.52, R) >droWil1.scaffold_180772 6499930 101 - 8906247 ----CUCAGAGCACUCGUCAGCACACUCAUUAUUAGGCUCAAGUGCAAAUUAGUGUGUUUGUUAGUGGGUUAUUAAGUUGGCCUGCCCACUUUUGCACACCCUUC---- ----......(((((.(..(((..............)))).)))))......((((((.....(((((((..............)))))))...)))))).....---- ( -25.58, z-score = -0.72, R) >droVir3.scaffold_12723 3833544 94 + 5802038 --UUCGAGGCACACUCGUCAGCGCACUCAUUAUUAGGCUCAAGUGCAAAUUAGUGUGUUUGUUAGUGGGUUAUUAGGUUCU-CCUGAGCCGCCUUAU------------ --...(((((..(((((..((((.((.(((((....((......))....))))).)).))))..))))).....(((((.-...))))))))))..------------ ( -31.40, z-score = -3.06, R) >droMoj3.scaffold_6500 9363752 99 - 32352404 UUCGCAUAGCACACUCGUCAGCGUACUCAUUAUUAGGCUCAAGUGCAAAUUAGUGUGUUUGUUAGUAGGUUAUUAAGUUUUGCCCAAGCCCCUCUGCGA---------- .(((((((((..(((....((((.((.(((((....((......))....))))).)).)))))))..)))))..((....((....))....))))))---------- ( -18.40, z-score = 0.65, R) >droGri2.scaffold_15252 8157422 94 + 17193109 --UUCGUGGCACACUCGUCAGCGCACUCAUUAUUAGGCUCAAGUGCAAAUUAGUGUGUUUGUUAGUAGGUUAUUAAGUUCU-CCUGAGCCUCCUUAC------------ --...(((((..(((..(.((((((((.(((.....((......)).))).)))))))).)..)))..)))))...((((.-...))))........------------ ( -18.90, z-score = 0.10, R) >consensus _____GUCGGCCACUCGUCAGCACACUCAUUAUUAGGCUCAAGUGCAAAUUAGUGUGUUUGCUAGUGGGUUAUUAAGUUCUCCCCCCGACACUCAGC____________ ............(((((..((((.((.(((((....((......))....))))).)).))))..)))))....................................... (-17.06 = -16.81 + -0.25)

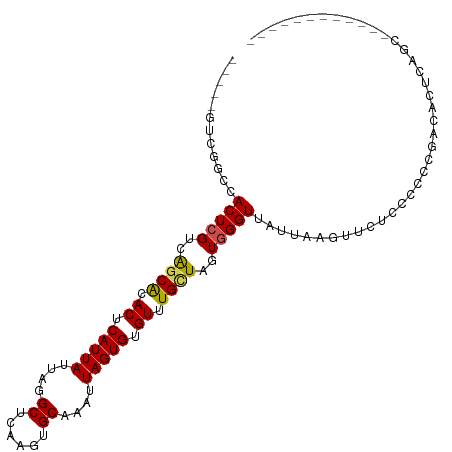
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:04 2011