
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,985,191 – 15,985,285 |
| Length | 94 |
| Max. P | 0.712528 |

| Location | 15,985,191 – 15,985,285 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 66.50 |
| Shannon entropy | 0.63611 |
| G+C content | 0.45993 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -7.63 |
| Energy contribution | -7.63 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
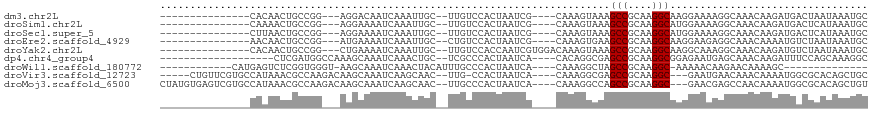
>dm3.chr2L 15985191 94 - 23011544 ---------------CACAACUGCCGG---AGGACAAUCAAAUUGC--UUGUCCACUAAUCG----CAAAGUAAAGCCGCAAGGCAAGGAAAAGGCAAACAAGAUGACUAAUAAAUGC ---------------......((((((---.((((((.(.....).--)))))).)).....----.........(((....)))........))))..................... ( -22.90, z-score = -2.13, R) >droSim1.chr2L 15699741 94 - 22036055 ---------------CAAAACUGCCGG---AGGAAAAUCAAAUUGC--UUGUCCACUAAUCG----CAAAGUAAAGCCGCAAGGCAUGGAAAAGGCAAACAAGAUGACUCAUAAAUGC ---------------...........(---((....(((...((((--((.((((((.....----...)))...(((....)))..)))..))))))....)))..)))........ ( -21.40, z-score = -1.47, R) >droSec1.super_5 4718634 94 - 5866729 ---------------CUUAACUGCCGG---AGGAAAAUCAAAUUGC--UUGUCCACUAAUCG----CAAAGUAAAGCCGCAAGGCAUGGAAAAGGCAAACAAGAUGACUCAUAAAUGC ---------------...........(---((....(((...((((--((.((((((.....----...)))...(((....)))..)))..))))))....)))..)))........ ( -21.40, z-score = -1.23, R) >droEre2.scaffold_4929 7682453 94 - 26641161 ---------------AACAACUGCCGG---AUGAAAAUCAAAUUGC--CUGUCCACUAAUCG----CAAAGUGAAGCCGCAAGGCAAGGAAGAGGCAAACAAAAUGUCUAAUAAAUGC ---------------.(((.......(---((....)))...((((--((.(((.....(((----(...)))).(((....)))..)))..))))))......)))........... ( -25.20, z-score = -2.74, R) >droYak2.chr2L 2975749 98 + 22324452 ---------------CACAACUGCCGG---CUGAAAAUCAAAUUGC--UUGUCCACCAAUCGUGGACAAAGUAAAGCCGCAAGGCAAGGCAAAGGCAAACAAGAUGUCUAAUAAAUGC ---------------......((((.(---((..........((((--((((((((.....)))))).)))))).(((....)))..)))...))))..................... ( -32.10, z-score = -3.90, R) >dp4.chr4_group4 4825959 93 - 6586962 -------------------CUCGAUGGCCAAAGCAAAUCAAACUGC--UCGCCCACUAAUCA----CACAGGCGAGCCGCAAGGCGGAGAAUGAGCAAACAAGAUUUCCAGCAAAGGC -------------------.......(((...(((((((....(((--(((((.........----....)))..(((....))).......))))).....)))))...))...))) ( -24.32, z-score = -0.92, R) >droWil1.scaffold_180772 6497566 86 - 8906247 ------------CAUGAGUCUCGGUGGGU-AAGCAAAUCAAACUACAUUUGCCCACUAAUCA----CAAAGGCUAGCCGCAAGGC-AAAAACAAGAACAAAAGC-------------- ------------....(((((..((((((-..((((((........))))))..)))...))----)..))))).(((....)))-..................-------------- ( -22.30, z-score = -2.90, R) >droVir3.scaffold_12723 3831368 103 + 5802038 -----CUGUUCGUGCCAUAAACGCCAAGACAAGCAAAUCAAGCAAC--UUG-CCACUAAUCA----CAAAGGCGAGCCGCAAGGC---GAAUGAACAAACAAAAUGGCGCACAGCUGC -----((((..(((((((...((((..((........))..((...--..)-).........----....)))).(((....)))---...............))))))))))).... ( -28.80, z-score = -1.78, R) >droMoj3.scaffold_6500 9361643 109 - 32352404 CUAUGUGAGUCGUGCCAUAAACGCCAAGACAAGCAAAUCAAGCAAC--UUGCCCACUAAUCA----CAAAGGCCAGCCGCAAGGC---GAACGAGCCAACAAAAUGGCGCACAGCUGU ...((((..(((((((......((........)).......((...--..))..........----....)))..(((....)))---..))))((((......)))).))))..... ( -27.10, z-score = -0.40, R) >consensus _______________CACAACUGCCGG___AAGAAAAUCAAAUUGC__UUGUCCACUAAUCG____CAAAGUAAAGCCGCAAGGCAAGGAAAAGGCAAACAAGAUGACUAAUAAAUGC ...........................................................................(((....)))................................. ( -7.63 = -7.63 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:03 2011