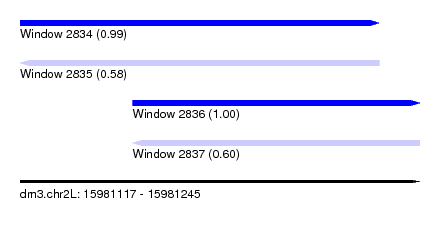
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,981,117 – 15,981,245 |
| Length | 128 |
| Max. P | 0.996317 |

| Location | 15,981,117 – 15,981,232 |
|---|---|
| Length | 115 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 76.91 |
| Shannon entropy | 0.50687 |
| G+C content | 0.35652 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -7.20 |
| Energy contribution | -8.34 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
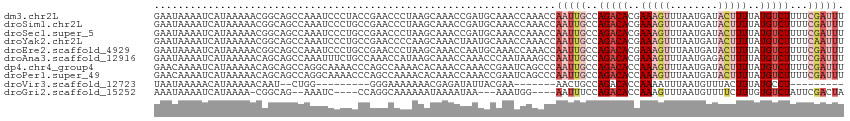
>dm3.chr2L 15981117 115 + 23011544 GAAUAAAAUCAUAAAAACGGCAGCCAAAUCCCUACCGAACCCUAAGCAAACCGAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUUU .....(((((........(((((.....((......)).......(((......)))............)))))(((((..(((((........)))))..)))))....))))) ( -20.70, z-score = -3.54, R) >droSim1.chr2L 15695027 115 + 22036055 GAAUAAAAUCAUAAAAACGGCAGCCAAAUCCCUGCCGAACCCUAAGCAAACCGAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUUU .....(((((.......((((((........))))))........(((......))).................(((((..(((((........)))))..)))))....))))) ( -24.70, z-score = -4.39, R) >droSec1.super_5 4714378 115 + 5866729 GAAUAAAAUCAUAAAAACGGCAGCCAAAUCCCUGCCGAACCCUAAGCAAACCGAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUUU .....(((((.......((((((........))))))........(((......))).................(((((..(((((........)))))..)))))....))))) ( -24.70, z-score = -4.39, R) >droYak2.chr2L 2970749 115 - 22324452 GAAUAAAAUCAUAAAAACGGCAGCCAAAUCCCUGCCGAACCCCAAGCAAACUAAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCAAUUU .................((((((........))))))........(((......)))..........(((((..(((((..(((((........)))))..)))))...))))). ( -24.60, z-score = -4.89, R) >droEre2.scaffold_4929 7678219 115 + 26641161 GAAUAAAAUCAUAAAAACGGCAGCCAAAUCCCUGCCGAACCCUAAGCAAACCAAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUUU .....(((((.......((((((........))))))........(((......))).................(((((..(((((........)))))..)))))....))))) ( -24.70, z-score = -4.66, R) >droAna3.scaffold_12916 15787835 115 - 16180835 GAAUAAAAUCAUAAAAACAGCAGCCAAAUUUCUGCCAAACCAUAAGCAAACCAAACCCAAUAAAGCCAAUUGCCAGACACGAAAGUUUAAUGAGACUUUUAUGUCUUUUCGAUUU .....(((((.........((((........))))..........((((....................)))).(((((..(((((((....)))))))..)))))....))))) ( -18.05, z-score = -2.80, R) >dp4.chr4_group4 4819933 115 + 6586962 GAACAAAAUCAUAAAAACAGCAGCCAGGCAAAACCCAGCCAAAACACAAACCAAACCGAAUCAGCCCAAUUGCCAGACACCAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUUU .....(((((................(((........))).................(((...((......)).(((((..(((((........)))))..))))).)))))))) ( -15.30, z-score = -1.84, R) >droPer1.super_49 257218 115 - 569410 GAACAAAAUCAUAAAAACAGCAGCCAGGCAAAACCCAGCCAAAACACAAACCAAACCGAAUCAGCCCAAUUGCCAGACACCAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUUU .....(((((................(((........))).................(((...((......)).(((((..(((((........)))))..))))).)))))))) ( -15.30, z-score = -1.84, R) >droVir3.scaffold_12723 1977059 88 + 5802038 UAAUAAAAACAUAAAAACAAU--CUGG---------GGGAAAAAAACGAGAUAUUACGAA-------AACUGCCAGACACCAAAAUUUAAUGUUUACUUUAUGCCU--------- .........((((((.....(--((((---------.((.......((........))..-------..)).))))).....((((.....))))..))))))...--------- ( -9.00, z-score = -0.46, R) >droGri2.scaffold_15252 8151148 101 - 17193109 AAAUAAAAUCAUAAAA-CGGCAG--AAAUC----CCAGGCAAAAAAUAAAAUAA---AAAUGG----AAUUUCCAGACACCAAAGUUUAAUGUUUUCUUUGUGUCUAUUCGACUA ................-(((..(--((((.----(((.................---...)))----.))))).((((((.((((...........))))))))))..))).... ( -14.15, z-score = -0.82, R) >consensus GAAUAAAAUCAUAAAAACGGCAGCCAAAUCCCUGCCGAACCAUAAGCAAACCAAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUUU ...................................................................(((((..(((((..(((((........)))))..)))))...))))). ( -7.20 = -8.34 + 1.14)

| Location | 15,981,117 – 15,981,232 |
|---|---|
| Length | 115 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.91 |
| Shannon entropy | 0.50687 |
| G+C content | 0.35652 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -10.88 |
| Energy contribution | -11.63 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.581449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
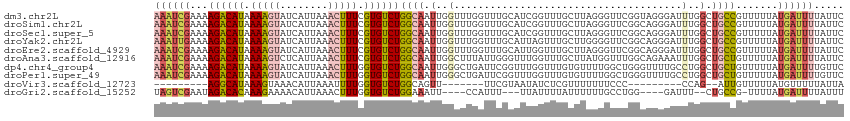
>dm3.chr2L 15981117 115 - 23011544 AAAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUCGGUUUGCUUAGGGUUCGGUAGGGAUUUGGCUGCCGUUUUUAUGAUUUUAUUC (((((((..(((((..(((((........)))))..))))).((((..........)))).))))))).....(((..((((((........))))))..)))............ ( -27.40, z-score = -1.56, R) >droSim1.chr2L 15695027 115 - 22036055 AAAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUCGGUUUGCUUAGGGUUCGGCAGGGAUUUGGCUGCCGUUUUUAUGAUUUUAUUC (((((((..(((((..(((((........)))))..))))).((((..........)))).))))))).....(((..((((((........))))))..)))............ ( -29.40, z-score = -1.92, R) >droSec1.super_5 4714378 115 - 5866729 AAAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUCGGUUUGCUUAGGGUUCGGCAGGGAUUUGGCUGCCGUUUUUAUGAUUUUAUUC (((((((..(((((..(((((........)))))..))))).((((..........)))).))))))).....(((..((((((........))))))..)))............ ( -29.40, z-score = -1.92, R) >droYak2.chr2L 2970749 115 + 22324452 AAAUUGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUUAGUUUGCUUGGGGUUCGGCAGGGAUUUGGCUGCCGUUUUUAUGAUUUUAUUC .(((((...(((((..(((((........)))))..)))))..)))))..........(((......)))...(((..((((((........))))))..)))............ ( -27.70, z-score = -1.50, R) >droEre2.scaffold_4929 7678219 115 - 26641161 AAAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUUGGUUUGCUUAGGGUUCGGCAGGGAUUUGGCUGCCGUUUUUAUGAUUUUAUUC (((((((..(((((..(((((........)))))..))))).((((..........)))).))))))).....(((..((((((........))))))..)))............ ( -27.30, z-score = -1.29, R) >droAna3.scaffold_12916 15787835 115 + 16180835 AAAUCGAAAAGACAUAAAAGUCUCAUUAAACUUUCGUGUCUGGCAAUUGGCUUUAUUGGGUUUGGUUUGCUUAUGGUUUGGCAGAAAUUUGGCUGCUGUUUUUAUGAUUUUAUUC ((((((...(((((..(((((........)))))..)))))(((((..(((((....)))))....)))))..))))))(((((........))))).................. ( -24.30, z-score = -1.06, R) >dp4.chr4_group4 4819933 115 - 6586962 AAAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUGGUGUCUGGCAAUUGGGCUGAUUCGGUUUGGUUUGUGUUUUGGCUGGGUUUUGCCUGGCUGCUGUUUUUAUGAUUUUGUUC ((((((...((((((.(((((........))))).))))))((((.(..(((.(((((((((..(.......)..)))))))))..)))..).)))).......))))))..... ( -32.30, z-score = -3.07, R) >droPer1.super_49 257218 115 + 569410 AAAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUGGUGUCUGGCAAUUGGGCUGAUUCGGUUUGGUUUGUGUUUUGGCUGGGUUUUGCCUGGCUGCUGUUUUUAUGAUUUUGUUC ((((((...((((((.(((((........))))).))))))((((.(..(((.(((((((((..(.......)..)))))))))..)))..).)))).......))))))..... ( -32.30, z-score = -3.07, R) >droVir3.scaffold_12723 1977059 88 - 5802038 ---------AGGCAUAAAGUAAACAUUAAAUUUUGGUGUCUGGCAGUU-------UUCGUAAUAUCUCGUUUUUUUCCC---------CCAG--AUUGUUUUUAUGUUUUUAUUA ---------((((((((((....(((((.....)))))(((((..(..-------..((........)).......)..---------))))--)....))))))))))...... ( -11.40, z-score = -0.42, R) >droGri2.scaffold_15252 8151148 101 + 17193109 UAGUCGAAUAGACACAAAGAAAACAUUAAACUUUGGUGUCUGGAAAUU----CCAUUU---UUAUUUUAUUUUUUGCCUGG----GAUUU--CUGCCG-UUUUAUGAUUUUAUUU .(((((...((((((((((...........)))).))))))(((((((----(((...---.................)))----)))))--))....-.....)))))...... ( -18.75, z-score = -1.10, R) >consensus AAAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUUGGUUUGCUUAGGGUUCGGCAGGGAUUUGGCUGCCGUUUUUAUGAUUUUAUUC ((((((...((((((.(((((........))))).))))))((((.(..(......................................)..).)))).......))))))..... (-10.88 = -11.63 + 0.75)
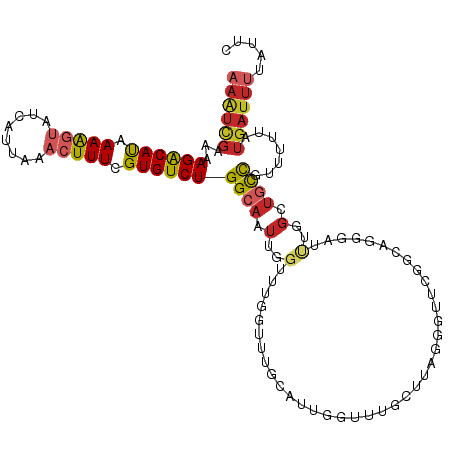
| Location | 15,981,153 – 15,981,245 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 79.98 |
| Shannon entropy | 0.38487 |
| G+C content | 0.33677 |
| Mean single sequence MFE | -14.14 |
| Consensus MFE | -9.72 |
| Energy contribution | -9.97 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.996317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15981153 92 + 23011544 GAACCCUAAGCAAACCGAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUU---UAACAACUUCUCUA----------- .........(((......)))..........(((((..(((((..(((((........)))))..)))))...)))))---..............----------- ( -14.30, z-score = -2.96, R) >droSim1.chr2L 15695063 92 + 22036055 GAACCCUAAGCAAACCGAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUU---UAACAACUUCUCUA----------- .........(((......)))..........(((((..(((((..(((((........)))))..)))))...)))))---..............----------- ( -14.30, z-score = -2.96, R) >droSec1.super_5 4714414 92 + 5866729 GAACCCUAAGCAAACCGAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUU---UAACAACUUCUCUA----------- .........(((......)))..........(((((..(((((..(((((........)))))..)))))...)))))---..............----------- ( -14.30, z-score = -2.96, R) >droYak2.chr2L 2970785 92 - 22324452 GAACCCCAAGCAAACUAAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCAAUU---UAACAACUUCUCUA----------- .........(((......)))..........(((((..(((((..(((((........)))))..)))))...)))))---..............----------- ( -14.60, z-score = -3.58, R) >droEre2.scaffold_4929 7678255 92 + 26641161 GAACCCUAAGCAAACCAAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUU---UAACAACUUCUCUA----------- .........(((......)))..........(((((..(((((..(((((........)))))..)))))...)))))---..............----------- ( -14.30, z-score = -3.21, R) >droAna3.scaffold_12916 15787871 92 - 16180835 AAACCAUAAGCAAACCAAACCCAAUAAAGCCAAUUGCCAGACACGAAAGUUUAAUGAGACUUUUAUGUCUUUUCGAUU---UAACAGCUUCUCUA----------- ..........................((((.(((((..(((((..(((((((....)))))))..)))))...)))))---.....)))).....----------- ( -15.40, z-score = -2.87, R) >dp4.chr4_group4 4819973 88 + 6586962 ----AAAACACAAACCAAACCGAAUCAGCCCAAUUGCCAGACACCAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUU---UAACAACUUUUCUA----------- ----.................(((((.((......)).(((((..(((((........)))))..)))))....))))---).............----------- ( -10.90, z-score = -2.35, R) >droPer1.super_49 257258 88 - 569410 ----AAAACACAAACCAAACCGAAUCAGCCCAAUUGCCAGACACCAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUU---UAACAACUUUUCUA----------- ----.................(((((.((......)).(((((..(((((........)))))..)))))....))))---).............----------- ( -10.90, z-score = -2.35, R) >droGri2.scaffold_15252 8151181 95 - 17193109 ---------AAAAAAUAAAAUAAA--AAUGGAAUUUCCAGACACCAAAGUUUAAUGUUUUCUUUGUGUCUAUUCGACUAACUGAAAAUUUCGAGAAAUUUCAUCAG ---------...............--.((((((((((.((((((.((((...........))))))))))..((((.............))))))))))))))... ( -18.22, z-score = -2.33, R) >consensus GAACCCUAAGCAAACCAAUGCAAACCAAACCAAUUGCCAGACACGAAAGUUUAAUGAUACUUUUAUGUCUUUUCGAUU___UAACAACUUCUCUA___________ ...............................(((((..(((((..(((((........)))))..)))))...)))))............................ ( -9.72 = -9.97 + 0.25)


| Location | 15,981,153 – 15,981,245 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.98 |
| Shannon entropy | 0.38487 |
| G+C content | 0.33677 |
| Mean single sequence MFE | -19.42 |
| Consensus MFE | -11.29 |
| Energy contribution | -11.03 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15981153 92 - 23011544 -----------UAGAGAAGUUGUUA---AAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUCGGUUUGCUUAGGGUUC -----------....(((.((..((---((((((..(((((..(((((........)))))..))))).((((..........)))).))))))))....)).))) ( -20.70, z-score = -1.96, R) >droSim1.chr2L 15695063 92 - 22036055 -----------UAGAGAAGUUGUUA---AAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUCGGUUUGCUUAGGGUUC -----------....(((.((..((---((((((..(((((..(((((........)))))..))))).((((..........)))).))))))))....)).))) ( -20.70, z-score = -1.96, R) >droSec1.super_5 4714414 92 - 5866729 -----------UAGAGAAGUUGUUA---AAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUCGGUUUGCUUAGGGUUC -----------....(((.((..((---((((((..(((((..(((((........)))))..))))).((((..........)))).))))))))....)).))) ( -20.70, z-score = -1.96, R) >droYak2.chr2L 2970785 92 + 22324452 -----------UAGAGAAGUUGUUA---AAUUGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUUAGUUUGCUUGGGGUUC -----------.....(((..(...---(((((...(((((..(((((........)))))..)))))..)))))....)..)))(((......)))......... ( -17.50, z-score = -0.79, R) >droEre2.scaffold_4929 7678255 92 - 26641161 -----------UAGAGAAGUUGUUA---AAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUUGGUUUGCUUAGGGUUC -----------....(((.((..((---((((((..(((((..(((((........)))))..))))).((((..........)))).))))))))....)).))) ( -18.60, z-score = -1.20, R) >droAna3.scaffold_12916 15787871 92 + 16180835 -----------UAGAGAAGCUGUUA---AAUCGAAAAGACAUAAAAGUCUCAUUAAACUUUCGUGUCUGGCAAUUGGCUUUAUUGGGUUUGGUUUGCUUAUGGUUU -----------.....((((.(..(---((((.((.(((((..(((((........)))))..)))))(((.....)))...)).)))))..)..))))....... ( -20.00, z-score = -1.21, R) >dp4.chr4_group4 4819973 88 - 6586962 -----------UAGAAAAGUUGUUA---AAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUGGUGUCUGGCAAUUGGGCUGAUUCGGUUUGGUUUGUGUUUU---- -----------...........(((---(((((((.((((((.(((((........))))).))))))(((......)))..))))))))))..........---- ( -20.10, z-score = -2.44, R) >droPer1.super_49 257258 88 + 569410 -----------UAGAAAAGUUGUUA---AAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUGGUGUCUGGCAAUUGGGCUGAUUCGGUUUGGUUUGUGUUUU---- -----------...........(((---(((((((.((((((.(((((........))))).))))))(((......)))..))))))))))..........---- ( -20.10, z-score = -2.44, R) >droGri2.scaffold_15252 8151181 95 + 17193109 CUGAUGAAAUUUCUCGAAAUUUUCAGUUAGUCGAAUAGACACAAAGAAAACAUUAAACUUUGGUGUCUGGAAAUUCCAUU--UUUAUUUUAUUUUUU--------- ..((((.((((((((((.............)))).(((((((((((...........)))).))))))))))))).))))--...............--------- ( -16.42, z-score = -0.68, R) >consensus ___________UAGAGAAGUUGUUA___AAUCGAAAAGACAUAAAAGUAUCAUUAAACUUUCGUGUCUGGCAAUUGGUUUGGUUUGCAUUGGUUUGCUUAGGGUUC .................(((((((............((((((.(((((........))))).)))))))))))))............................... (-11.29 = -11.03 + -0.25)
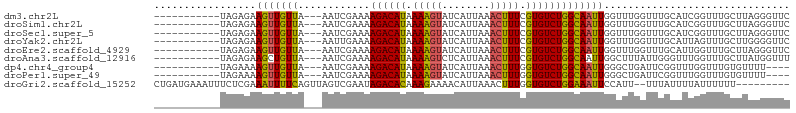
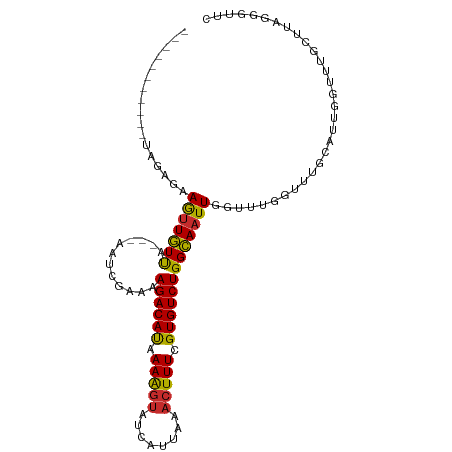
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:44:02 2011