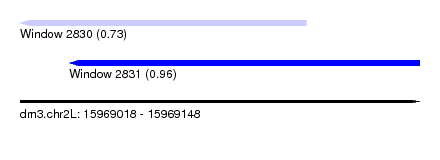
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,969,018 – 15,969,148 |
| Length | 130 |
| Max. P | 0.964555 |

| Location | 15,969,018 – 15,969,111 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 91.64 |
| Shannon entropy | 0.17058 |
| G+C content | 0.43659 |
| Mean single sequence MFE | -19.03 |
| Consensus MFE | -18.59 |
| Energy contribution | -17.79 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
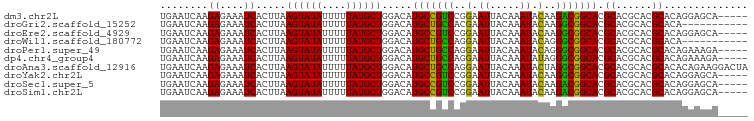
>dm3.chr2L 15969018 93 - 23011544 UGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGUCCGGAAUUACAAAUACAAGACGGCACGCACGCACGCACAGGAGCA----- ........((....)).....((((((....))))))(..(.(((((((..(.((.....)).)..))))))).)..)....((......)).----- ( -19.00, z-score = -0.98, R) >droGri2.scaffold_15252 8136564 87 + 17193109 UGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCACGAAUUACAAAUACAAGGCGGCACGCACGCACGCACA----------- ........((....)).....((((((....)))))).....(((((((.................))))))).((......))...----------- ( -17.73, z-score = -1.05, R) >droEre2.scaffold_4929 7666748 93 - 26641161 UGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGUCCGGAAUUACAAAUACAAGGCGGCACGCACGCACGCACAGGAGCA----- ........((....)).(((..((.(((((.((..((((((.....))))))...)).)))))....(((((......)).)))))..)))..----- ( -19.90, z-score = -0.72, R) >droWil1.scaffold_180772 6473332 87 - 8906247 UGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUACAGGGCGGCACGCACGCACGCACA----------- ........((....)).....((((((....)))))).....(((((((..(.((.....)).)..))))))).((......))...----------- ( -18.70, z-score = -1.04, R) >droPer1.super_49 232369 93 + 569410 UGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUACAGGGCGGCACGCACGCACGCACAGAAAGA----- ........((....)).(((.((((((....))))))(..(.(((((((..(.((.....)).)..))))))).)..)...........))).----- ( -18.80, z-score = -0.93, R) >dp4.chr4_group4 4795610 93 - 6586962 UGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUAUAGGGCGGCACGCACGCACGCACAGAAAGA----- ........((....)).(((.((((((....))))))(..(.(((((((.................))))))).)..)...........))).----- ( -17.73, z-score = -0.51, R) >droAna3.scaffold_12916 15775094 98 + 16180835 UGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUACUAGGCGGCACGCACGCACGCACACAGAAGGACUA ........((....)).....((((((....))))))(..(.(((((((((............)).))))))).((......))........)..).. ( -20.50, z-score = -1.33, R) >droYak2.chr2L 2956416 93 + 22324452 UGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGUCCGGAAUUACAAAUACAAGGCGGCACGCACGCACGCACAGGAGCA----- ........((....)).(((..((.(((((.((..((((((.....))))))...)).)))))....(((((......)).)))))..)))..----- ( -19.90, z-score = -0.72, R) >droSec1.super_5 4702478 93 - 5866729 UGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGUCCGGAAUUACAAAUACAAGACGGCACGCACGCACGCACAGGAGCA----- ........((....)).....((((((....))))))(..(.(((((((..(.((.....)).)..))))))).)..)....((......)).----- ( -19.00, z-score = -0.98, R) >droSim1.chr2L 15682146 93 - 22036055 UGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGUCCGGAAUUACAAAUACAAGACGGCACGCACGCACGCACAGGAGCA----- ........((....)).....((((((....))))))(..(.(((((((..(.((.....)).)..))))))).)..)....((......)).----- ( -19.00, z-score = -0.98, R) >consensus UGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGCCAGGAAUUACAAAUACAAGGCGGCACGCACGCACGCACAGGAGCA_____ ........((....)).....((((((....)))))).....(((((((..(.((.....)).)..))))))).((......)).............. (-18.59 = -17.79 + -0.80)

| Location | 15,969,034 – 15,969,148 |
|---|---|
| Length | 114 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.44 |
| Shannon entropy | 0.22787 |
| G+C content | 0.39501 |
| Mean single sequence MFE | -28.69 |
| Consensus MFE | -21.98 |
| Energy contribution | -21.39 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
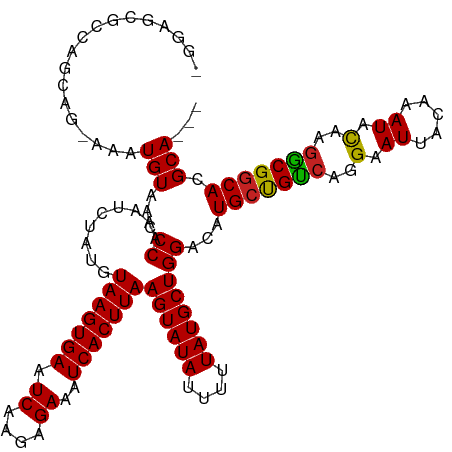
>dm3.chr2L 15969034 114 - 23011544 -GGAGCGCCAGCAG-AAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGUCCGGAAUUACAAAUACAAGACGGCACGCA--- -...((((((((((-(((((((.............(((((((.((....))..)))))))..))))))))..))))))...(((((((..(.((.....)).)..)))))))))).--- ( -33.26, z-score = -3.85, R) >droGri2.scaffold_15252 8136574 103 + 17193109 -------------AGAAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCACGAAUUACAAAUACAAGGCGGCACGCA--- -------------.....(((...((.........(((((((.((....))..)))))))((((((....))))))))...(((((((.................))))))).)))--- ( -22.43, z-score = -1.64, R) >droVir3.scaffold_12723 3805606 110 + 5802038 ---------AGAACAAAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGUCAGGAAUUACAAAUACAACGCGACACGCAGGC ---------.........(((((.((.........(((((((.((....))..)))))))((((((....)))))).(((.....))).))..)))))........(((...))).... ( -18.70, z-score = -0.27, R) >droEre2.scaffold_4929 7666764 114 - 26641161 -GGAGCGCCAGCAG-AAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGUCCGGAAUUACAAAUACAAGGCGGCACGCA--- -...((((((((((-(((((((.............(((((((.((....))..)))))))..))))))))..))))))...(((((((..(.((.....)).)..)))))))))).--- ( -32.66, z-score = -3.03, R) >droWil1.scaffold_180772 6473342 116 - 8906247 GAAAAGGCAAAGGAGAAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUACAGGGCGGCACGCA--- ......((.........((((...((.........(((((((.((....))..)))))))((((((....))))))))))))((((((..(.((.....)).)..))))))..)).--- ( -25.70, z-score = -1.26, R) >droPer1.super_49 232385 114 + 569410 --GAAACAGAAACAGAAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUACAGGGCGGCACGCA--- --.........(((....)))...((.........(((((((.((....))..)))))))((((((....))))))))...(((((((..(.((.....)).)..)))))))....--- ( -25.40, z-score = -1.93, R) >dp4.chr4_group4 4795626 114 - 6586962 --GAAACAGAAACAGAAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUAUAGGGCGGCACGCA--- --.........(((....)))...((.........(((((((.((....))..)))))))((((((....))))))))...(((((((.................)))))))....--- ( -24.33, z-score = -1.61, R) >droAna3.scaffold_12916 15775115 115 + 16180835 -GGAGCGCUAGCAGAAAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGCCAGGAAUUACAAAUACUAGGCGGCACGCA--- -...((((((((((((((((((.............(((((((.((....))..)))))))..))))))))).))))))...(((((((((............)).)))))))))).--- ( -33.96, z-score = -3.28, R) >droYak2.chr2L 2956432 114 + 22324452 -GGAGCGCCAGCAG-AAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGUCCGGAAUUACAAAUACAAGGCGGCACGCA--- -...((((((((((-(((((((.............(((((((.((....))..)))))))..))))))))..))))))...(((((((..(.((.....)).)..)))))))))).--- ( -32.66, z-score = -3.03, R) >droSec1.super_5 4702494 114 - 5866729 -GGAGCGCCAGCAG-AAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGUCCGGAAUUACAAAUACAAGACGGCACGCA--- -...((((((((((-(((((((.............(((((((.((....))..)))))))..))))))))..))))))...(((((((..(.((.....)).)..)))))))))).--- ( -33.26, z-score = -3.85, R) >droSim1.chr2L 15682162 114 - 22036055 -GGAGCGCCAGCAG-AAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCCGUCCGGAAUUACAAAUACAAGACGGCACGCA--- -...((((((((((-(((((((.............(((((((.((....))..)))))))..))))))))..))))))...(((((((..(.((.....)).)..)))))))))).--- ( -33.26, z-score = -3.85, R) >consensus _GGAGCGCCAGCAG_AAAUGUAACCCAAAUCUAUGUAAGUGAAUCAAGAGAAAUCACUUAAGUAUAUUUUUAUGCUGGACAUGCUGUCAGGAAUUACAAAUACAAGGCGGCACGCA___ ..................(((...((.........(((((((.((....))..)))))))((((((....))))))))...(((((((..(.((.....)).)..))))))).)))... (-21.98 = -21.39 + -0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:57 2011