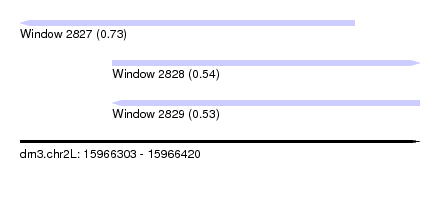
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,966,303 – 15,966,420 |
| Length | 117 |
| Max. P | 0.733703 |

| Location | 15,966,303 – 15,966,401 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.93 |
| Shannon entropy | 0.56875 |
| G+C content | 0.47211 |
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -14.15 |
| Energy contribution | -13.76 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.733703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15966303 98 - 23011544 AAAUCGGAAGCC-GAAAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCCGAA--CUCCAUUCCGAGAUCGGAAUAGCC----------------- .....(((...(-....)((((.(((.......)))..........(((.((((....))))..)))..))))..--.)))((((((....))))))....----------------- ( -29.20, z-score = -1.65, R) >droSim1.chr2L 15679386 98 - 22036055 AAAUCGGAAGCC-GAAAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCCGAA--CUCCAUUUCGAGAUCGGAAUAGCC----------------- ...((((...))-)).....((((((.......)))..........(((.((((....))))..)))...((((.--(((......))).))))....)))----------------- ( -26.90, z-score = -0.95, R) >droSec1.super_5 4699704 98 - 5866729 AAAUCGGAAGCC-GAAAGCGGGAGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCCGAA--CUCCAUUUCGAGAUCGGAAUAGCC----------------- .....((...((-((...((((.(((.......)))..........(((.((((....))))..)))..))))..--(((......))).)))).....))----------------- ( -27.10, z-score = -1.45, R) >droYak2.chr2L 2953763 97 + 22324452 AAAUCGGAAGCU-GAAAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGA-CCCGAA--CUCCAUUUCGAGAUCGGAAUAGCC----------------- .....(((.((.-....))(((.(((.......)))..........(((.((((....))))..))).-)))...--.)))((((((....))))))....----------------- ( -25.90, z-score = -0.92, R) >droEre2.scaffold_4929 7663852 98 - 26641161 AAAUCGGAAGCC-GAAAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCCGAA--CUCCAUUUCGAGAUCGGAAUAGCC----------------- ...((((...))-)).....((((((.......)))..........(((.((((....))))..)))...((((.--(((......))).))))....)))----------------- ( -26.90, z-score = -0.95, R) >droAna3.scaffold_12916 15772708 99 + 16180835 AGAUCGGAAGCCAAAAAACGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCUAAA--CUCCAUUUGGUGCUCAGGUUGGCC----------------- .........(((((.....(((((((.......))).........((((.((((....))))((((((.......--.))))))))))))))...))))).----------------- ( -30.40, z-score = -1.71, R) >dp4.chr4_group4 4792767 106 - 6586962 AAAUCGGAAGCC-AAAAACGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCUGAG--CCCCAUAGAGAGUGGGUGCCAUCCGAGGGGCG--------- ...(((((....-......(((.(((.......)))..........(((.((((....))))..))).)))((.(--((((((.....))))).)))))))))......--------- ( -33.10, z-score = -0.80, R) >droPer1.super_49 229528 106 + 569410 AAAUCGGAAGCC-AAAAACGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCUGAG--CCCCAUAGAGAGUGGGUGCCAUCCGAGGGGCG--------- ...(((((....-......(((.(((.......)))..........(((.((((....))))..))).)))((.(--((((((.....))))).)))))))))......--------- ( -33.10, z-score = -0.80, R) >droWil1.scaffold_180772 2438658 93 + 8906247 UAAUAGGAAGCCAAAAAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCUGAA--ACUCCUGUGAAGGAAGCA----------------------- ..((((((.((......))(((.(((.......)))..........(((.((((....))))..))).)))....--..))))))..........----------------------- ( -23.70, z-score = -0.96, R) >droMoj3.scaffold_6500 9327960 113 - 32352404 AAAUAGGAAGUC--AAAACGAGCGCAUAAAAAUUGCAUUAAAAUUGCUUUGCGGAUAUCCGCAAUGGAGUCCAU---UUUCGGCUUCUUACAACGUUUGCCUUCAGUUCCAUUUGCCG .....((((.(.--((..((((((..........(((.......))).((((((....)))))).((((((...---....))))))......))))))..)).).))))........ ( -26.40, z-score = -1.00, R) >droVir3.scaffold_12723 3801336 113 + 5802038 AAAUAGGAAGCC--AAAACGAGCGCAUAAAAAUUGCAUUAAAAUAGCCGUGCGGAUAUCCGCAAUGGACUCCAUU---UUCGAGGUUUGUUGGAUUUUUCCUCUUCUUUUUAUUUGUU ((((((((((..--..((((((((((.......)))..........((((((((....)))).)))).(((....---...))))))))))(((....)))....))))))))))... ( -29.50, z-score = -1.96, R) >droGri2.scaffold_15252 8132794 116 + 17193109 AAAUAGGAAGCC--AAAACGAGCGCAUAAAAAUUGCAUUAAAAAUGCUGUGCGGAUAUCCGCAAUGGACUCCAUUUCCUUCAGAGCCAAUUCAGAUUUUCAGUUUCAGUUUUUUGGUU ........((((--(((..(((((((.......)))(((.(((((.((((((((....))))).(((.(((...........))))))...)))))))).)))....))))))))))) ( -26.30, z-score = -0.97, R) >consensus AAAUCGGAAGCC_GAAAACGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCCGAA__CUCCAUUUCGAGAUCGGAAUAGCC_________________ ...................(((.(((.......)))..........(((.((((....))))..))).)))............................................... (-14.15 = -13.76 + -0.39)

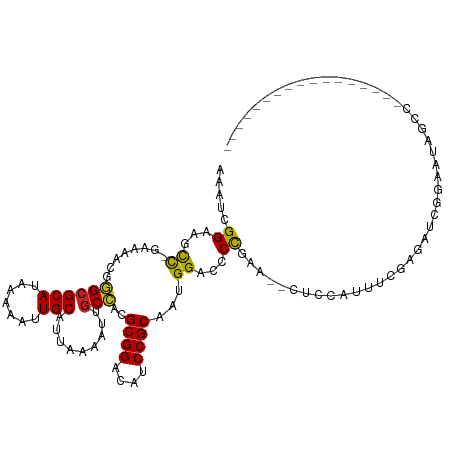

| Location | 15,966,330 – 15,966,420 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Shannon entropy | 0.41882 |
| G+C content | 0.43245 |
| Mean single sequence MFE | -21.34 |
| Consensus MFE | -17.07 |
| Energy contribution | -16.32 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.28 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.543873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15966330 90 + 23011544 --------GGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGCUUUC-GGCUUCCGAUUUUUU------AUUUAACCCCCCCGUU-------- --------(((((((((.((((....))))))))..........(((.......)))(((.......-)))............------.....)))))......-------- ( -23.10, z-score = -0.14, R) >droSim1.chr2L 15679413 89 + 22036055 --------GGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGCUUUC-GGCUUCCGAUUUUUU------AUUUAACCCCCC-GUU-------- --------(((((((((.((((....))))))))..........(((.......)))(((.......-)))............------.....)))))..-...-------- ( -23.10, z-score = -0.19, R) >droSec1.super_5 4699731 89 + 5866729 --------GGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCUCCCGCUUUC-GGCUUCCGAUUUUUU------AUUUAACCCCCC-GUU-------- --------(((((((((.((((....))))))))..........(((.......)))........((-((...))))......------.....)))))..-...-------- ( -22.10, z-score = -0.42, R) >droYak2.chr2L 2953790 88 - 22324452 ---------GGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGCUUUC-AGCUUCCGAUUUUUU------AUUUAACCCCCC-GUU-------- ---------((((((((.((((....))))))))..........(((.......)))))))((....-.))............------............-...-------- ( -19.20, z-score = -0.18, R) >droEre2.scaffold_4929 7663879 89 + 26641161 --------GGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGCUUUC-GGCUUCCGAUUUUUU------AUUUAACCCCCC-GUU-------- --------(((((((((.((((....))))))))..........(((.......)))(((.......-)))............------.....)))))..-...-------- ( -23.10, z-score = -0.19, R) >droAna3.scaffold_12916 15772735 99 - 16180835 --------AGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGUUUUUUGGCUUCCGAUCUUUU------AUUUCGUUCUCUCAUCCUCCCACC --------.(((.((((.((((....))))))))..........(((.......)))(((........)))............------..................)))... ( -19.70, z-score = -0.22, R) >droWil1.scaffold_180772 2438679 90 - 8906247 --------AGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGCUUUUUGGCUUCCUAUUAUAU------AUUCGGUUUUCUUUU--------- --------.((((((((.((((....))))))))..........(((.......)))))))(((....)))............------...............--------- ( -20.60, z-score = -0.43, R) >droMoj3.scaffold_6500 9327997 100 + 32352404 --GAAAAUGGACUCCAUUGCGGAUAUCCGCAAAGCAAUUUUAAUGCAAUUUUUAUGCGCUCGUUUU--GACUUCCUAUUUGCCCUGGCUACAAUAUUUUUUAUU--------- --.((((((..(....((((((....)))))).(((.......)))...........)..))))))--............((....))................--------- ( -17.40, z-score = -0.36, R) >droVir3.scaffold_12723 3801373 90 - 5802038 --GAAAAUGGAGUCCAUUGCGGAUAUCCGCACGGCUAUUUUAAUGCAAUUUUUAUGCGCUCGUUUU--GGCUUCCUAUUUUCU----------UAUAUUUUAUU--------- --((((((((((.(((..((((....))))(((((.........(((.......))))).)))..)--)).)))).)))))).----------...........--------- ( -21.80, z-score = -2.25, R) >droGri2.scaffold_15252 8132832 97 - 17193109 AAGGAAAUGGAGUCCAUUGCGGAUAUCCGCACAGCAUUUUUAAUGCAAUUUUUAUGCGCUCGUUUU--GGCUUCCUAUUUUCG-----UAUUUUAUUUUUUAUU--------- ..((((((((((.(((.(((((....)))))..((((.....))))...................)--)).)))).)))))).-----................--------- ( -23.30, z-score = -2.38, R) >consensus ________GGGGUCCAUUGCGGAUGUCCGCGUGGCAAUUUUAAUGCAAUUUUUAUGCGCCCGCUUUC_GGCUUCCGAUUUUUU______AUUUAACCCCCC_GUU________ .........((((.((((((((....)))))..(((.......))).......))).))))(((....))).......................................... (-17.07 = -16.32 + -0.75)
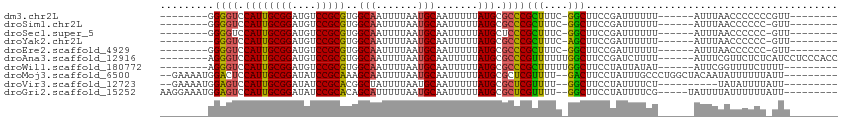
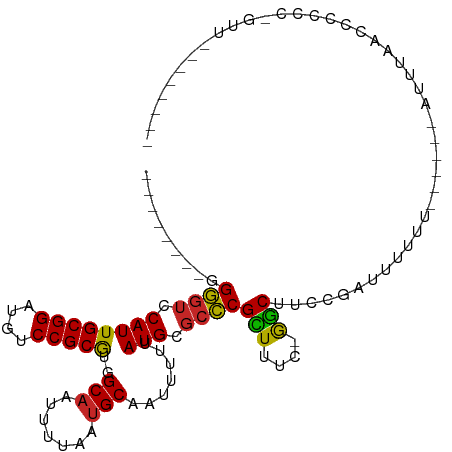
| Location | 15,966,330 – 15,966,420 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.95 |
| Shannon entropy | 0.41882 |
| G+C content | 0.43245 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -14.16 |
| Energy contribution | -13.64 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15966330 90 - 23011544 --------AACGGGGGGGUUAAAU------AAAAAAUCGGAAGCC-GAAAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCC-------- --------......((((((....------........((..(((-.......)))(((.......)))..........))..((((....))))....))))))-------- ( -26.00, z-score = -0.89, R) >droSim1.chr2L 15679413 89 - 22036055 --------AAC-GGGGGGUUAAAU------AAAAAAUCGGAAGCC-GAAAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCC-------- --------...-..((((((....------........((..(((-.......)))(((.......)))..........))..((((....))))....))))))-------- ( -26.00, z-score = -1.12, R) >droSec1.super_5 4699731 89 - 5866729 --------AAC-GGGGGGUUAAAU------AAAAAAUCGGAAGCC-GAAAGCGGGAGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCC-------- --------...-..((((((....------......((((...))-))..(((((.(((.......)))........((((....)).)))))))....))))))-------- ( -24.70, z-score = -1.19, R) >droYak2.chr2L 2953790 88 + 22324452 --------AAC-GGGGGGUUAAAU------AAAAAAUCGGAAGCU-GAAAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCC--------- --------...-...(((((....------........((..(((-.......)))(((.......)))..........))..((((....))))....)))))--------- ( -20.20, z-score = -0.07, R) >droEre2.scaffold_4929 7663879 89 - 26641161 --------AAC-GGGGGGUUAAAU------AAAAAAUCGGAAGCC-GAAAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCC-------- --------...-..((((((....------........((..(((-.......)))(((.......)))..........))..((((....))))....))))))-------- ( -26.00, z-score = -1.12, R) >droAna3.scaffold_12916 15772735 99 + 16180835 GGUGGGAGGAUGAGAGAACGAAAU------AAAAGAUCGGAAGCCAAAAAACGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCU-------- .((((.((..........(((...------......)))...(((........)))(((.......))).......)).))))((((....))))..........-------- ( -21.70, z-score = -0.52, R) >droWil1.scaffold_180772 2438679 90 + 8906247 ---------AAAAGAAAACCGAAU------AUAUAAUAGGAAGCCAAAAAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCU-------- ---------.........(((...------........((..(((........)))(((.......)))..........))..((((....))))..))).....-------- ( -18.90, z-score = -1.05, R) >droMoj3.scaffold_6500 9327997 100 - 32352404 ---------AAUAAAAAAUAUUGUAGCCAGGGCAAAUAGGAAGUC--AAAACGAGCGCAUAAAAAUUGCAUUAAAAUUGCUUUGCGGAUAUCCGCAAUGGAGUCCAUUUUC-- ---------................((....)).....(((..((--(........(((.......)))............((((((....)))))))))..)))......-- ( -21.10, z-score = -1.05, R) >droVir3.scaffold_12723 3801373 90 + 5802038 ---------AAUAAAAUAUA----------AGAAAAUAGGAAGCC--AAAACGAGCGCAUAAAAAUUGCAUUAAAAUAGCCGUGCGGAUAUCCGCAAUGGACUCCAUUUUC-- ---------...........----------.((((((.(((..((--(..(((.(((((.......)))((....)).)))))((((....))))..)))..)))))))))-- ( -22.80, z-score = -3.47, R) >droGri2.scaffold_15252 8132832 97 + 17193109 ---------AAUAAAAAAUAAAAUA-----CGAAAAUAGGAAGCC--AAAACGAGCGCAUAAAAAUUGCAUUAAAAAUGCUGUGCGGAUAUCCGCAAUGGACUCCAUUUCCUU ---------................-----........(((..((--(........(((.......))).............(((((....))))).)))..)))........ ( -19.30, z-score = -1.93, R) >consensus ________AAC_AGAGGAUUAAAU______AAAAAAUCGGAAGCC_GAAAGCGGGCGCAUAAAAAUUGCAUUAAAAUUGCCACGCGGACAUCCGCAAUGGACCCC________ ....................................................(((.(((.......)))..........(((.((((....))))..))).)))......... (-14.16 = -13.64 + -0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:55 2011