| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,964,123 – 15,964,214 |
| Length | 91 |
| Max. P | 0.524270 |

| Location | 15,964,123 – 15,964,214 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.66 |
| Shannon entropy | 0.49308 |
| G+C content | 0.43149 |
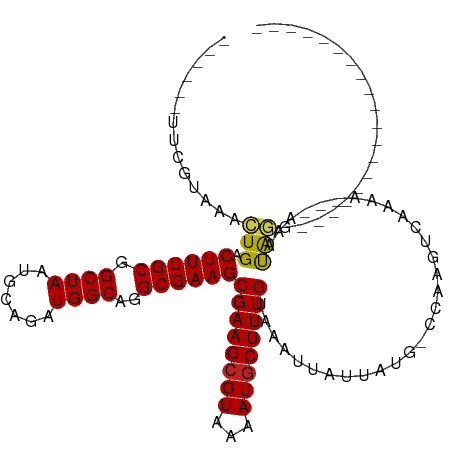
| Mean single sequence MFE | -24.29 |
| Consensus MFE | -16.48 |
| Energy contribution | -16.12 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524270 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
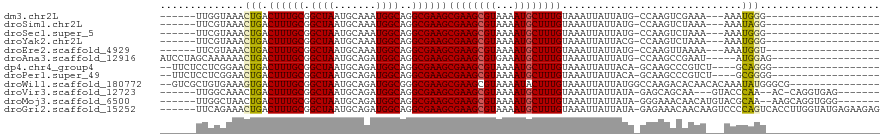
>dm3.chr2L 15964123 91 + 23011544 ------UUGGUAAACUGACUUUGCGGCUAAUGCAAAUGGCAGGCGAAGCGAAGCGUAAAAUGCUUUGUAAAUUAUUAUG-CCAAGUCGAAA---AAAUGGG------------------- ------((((((..(((..((((((.....))))))...))).....(((((((((...))))))))).........))-)))).......---.......------------------- ( -22.90, z-score = -1.21, R) >droSim1.chr2L 15677240 91 + 22036055 ------UUCGUAAACUGACUUUGCGGCUAAUGCAAAUGGCAGGCGAAGCGAAGCGUAAAAUGCUUUGUAAAUUAUUAUG-CCAAGUCUAAA---AAAUAGG------------------- ------..........(((((.((((((..(((.....))))))...(((((((((...))))))))).........))-).)))))....---.......------------------- ( -21.90, z-score = -1.17, R) >droSec1.super_5 4697559 91 + 5866729 ------UUCGUAAACUGACUUUGCGGCUAAUGCAAAUGGCAGGCGAAGCGAAGCGUAAAAUGCUUUGUAAAUUAUUAUG-CCAAGUCUAAA---AAAUGGG------------------- ------.((((.....(((((.((((((..(((.....))))))...(((((((((...))))))))).........))-).)))))....---..)))).------------------- ( -22.40, z-score = -1.19, R) >droYak2.chr2L 2951658 91 - 22324452 ------UUCGUAAACUGACUUUGCGGCUAAUGCAAAUGGCAGGCGAAGCGAAGCGUAAAAUGCUUUGUAAAUUAUUACG-CCAAGUCUAAA---AAAUGGG------------------- ------.((((.....(((((.((((((..(((.....))))))...(((((((((...))))))))).........))-).)))))....---..)))).------------------- ( -24.60, z-score = -1.75, R) >droEre2.scaffold_4929 7661720 91 + 26641161 ------UUCGUAAACUGACUUUGCGGCUAAUGCAAAUGGCAGGCGAAGCGAAGCGUAAAAUGCUUUGUAAAUUAUUAUG-CCAAGUUAAAA---AAAUGGU------------------- ------(((((...(((..((((((.....))))))...))))))))(((((((((...)))))))))..........(-(((........---...))))------------------- ( -21.10, z-score = -0.73, R) >droAna3.scaffold_12916 15761890 96 - 16180835 AUCCUAGCAAAAAACUGACUUUGCGGCUAAUGCAGAUGGCAGGCGAAGCGAAGCGUGAAAUGCUUUGUAAAUUAUUAUG-CCAAGCCGAAU-----AUGGAG------------------ .(((..(((((........)))))((((..(((.....)))(((((((((((((((...)))))))))......)).))-)).))))....-----..))).------------------ ( -26.80, z-score = -1.41, R) >dp4.chr4_group4 1796244 95 - 6586962 --UUCUCCUCGGAACUGACUUUGCGGCUAAUGCAGAUGGCAGGCGAAGCGAAGCGUAAAAUGCUUUGUAAAUUAUUACA-GCAAGCCCGUCU----GCAGGG------------------ --....(((((.((......)).)))....((((((((((..((.....(((((((...)))))))((((....)))).-))..).))))))----))))).------------------ ( -26.60, z-score = 0.01, R) >droPer1.super_49 341937 95 + 569410 --UUCUCCUCGGAACUGACUUUGCGGCUAAUGCAGAUGGCAGGCGAAGCGAAGCGUAAAAUGCUUUGUAAAUUAUUACA-GCAAGCCCGUCU----GCGGGG------------------ --....((((.........((((((.....))))))..(((((((..(((((((((...)))))))((((....)))).-....)).)))))----))))))------------------ ( -28.40, z-score = -0.35, R) >droWil1.scaffold_180772 6465303 103 + 8906247 --GUCGCUGUGAAAGUGACUUUGCGGCUAAUGCAGAUGGCGGGCGAAGCGAAGCGUAAAAUACUUUGUAAAUUAUUAUGGCCAAGACACAACACAAAUAUGGGCG--------------- --((((((.....))))))...(((((((.(((.....)))......((((((.........)))))).........)))))...........((....)).)).--------------- ( -20.80, z-score = 1.34, R) >droVir3.scaffold_12723 2004261 100 + 5802038 ------UUGGCAAACUGACUUUGCGGCUAAUGCAGAUGGCAGGCGAAGCGAAGCGUAAAAUGCUUUGUAAAUUAUUAUA-GAGCAGCAA---GUACCCAA--AC-CAGGUGAG------- ------...((.....(.((((((.(((..(((.....))))))...)))))))......((((((((((....)))))-)))))))..---.((((...--..-..))))..------- ( -25.70, z-score = -0.42, R) >droMoj3.scaffold_6500 23028391 104 - 32352404 ------UUGGCUAACUGACUUUGCGGCUAAUGCAGAUGGCAGGCGAAGCGAAGCGUAAAAUGCUUUGUAAAUUAUUAUA-GGGAAACAACAUGUACGCAA--AAGCAGGUGGG------- ------....(((.(((..(((((((((..(((.....))))))...(((((((((...))))))))).......((((-.(....)....)))))))))--)..))).))).------- ( -28.90, z-score = -1.78, R) >droGri2.scaffold_15252 9063248 113 + 17193109 ------UUCAGAAACUGACUUUGCGGCUAAUGCAGAUGGCAGGCGAAGCGAAGCGUAAAAUGCUUUGUAAAUUAUUAUA-GAGAAACAACAAGUCCCCAGUCACCUUGGUAUGAGAAGAG ------((((......((((((((.((((.......))))..)))))(((((((((...)))))))))...........-............))).((((.....))))..))))..... ( -21.40, z-score = 0.78, R) >consensus ______UUCGUAAACUGACUUUGCGGCUAAUGCAGAUGGCAGGCGAAGCGAAGCGUAAAAUGCUUUGUAAAUUAUUAUG_CCAAGUCAAAA___AAAUAGG___________________ ..............(((.((((((.((((.......))))..))))))((((((((...))))))))..............................))).................... (-16.48 = -16.12 + -0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:53 2011