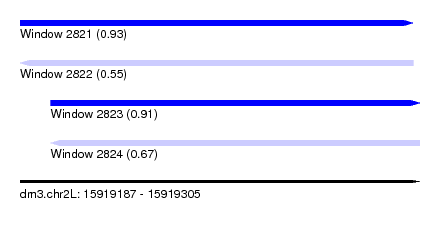
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,919,187 – 15,919,305 |
| Length | 118 |
| Max. P | 0.932972 |

| Location | 15,919,187 – 15,919,303 |
|---|---|
| Length | 116 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Shannon entropy | 0.47407 |
| G+C content | 0.44274 |
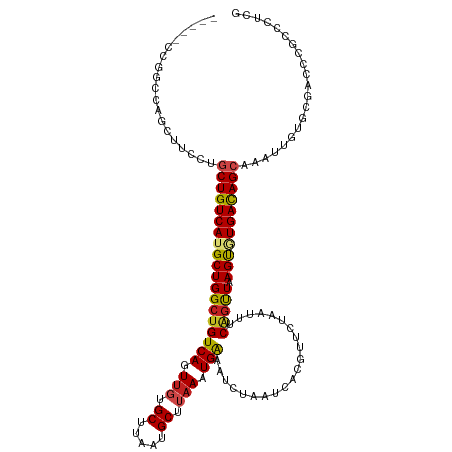
| Mean single sequence MFE | -28.36 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.59 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15919187 116 + 23011544 CAUGCCCGGCAAGCUCCCUGCUGUCAUGCUGGCUGUCAGUUUAGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUGUGACAGCGAAUUGUGCGACCCGCCC--- ......(((...((.(..(((((((((...(((((......)))))....((((((.((((((..............)))))).)))))))))))))))....).))...)))...--- ( -29.64, z-score = -1.15, R) >droSim1.chr2L 15631988 119 + 22036055 CAUGCCCGGGAAGCUCCCUGCUGUCAUGCUGGCUGUCAGUUGCGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUGUGACAGCGAAUUGUGCGACCCGCCCUUG ......((((..((.(..(((((((((((((((((..(((((.((.....)).....(((.......))).....))))).))))).))))))))))))....).))..))))...... ( -33.10, z-score = -1.55, R) >droSec1.super_5 4653254 119 + 5866729 CAUGCCGGGGAAGCUCCCUGCUGUCAUGCUGGCUGUCAGUUGCGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUGUGACAGCGAAUUGUGCGACCCGCCCUUG .....((((...((.(..(((((((((((((((((..(((((.((.....)).....(((.......))).....))))).))))).))))))))))))....).))..))))...... ( -32.80, z-score = -0.90, R) >droYak2.chr2L 2904908 119 - 22324452 CAUGCUCAGGAAGCUCUGUGCUGUCAUGCUGGCUGUCAGUUGCGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCGGUUAAGUGUGACAGCGAAUUGUGACCCCCCCCCCCA ...((.((((....)))).)).(((((..(.((((((....(((((((((.......(((.......)))((.........))))))))))))))))).)...)))))........... ( -27.30, z-score = -0.37, R) >droEre2.scaffold_4929 7618290 118 + 26641161 CAUGCCCGGGCAGCUCC-UGCUGUCAUGCUGGCUGUCAGUGGCGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUGUGACAGCGAAUCGUGUGACCCGCCCUCG .......(((((((...-.)))((((((((.((((((....(((((((((.......((((((..............))))))))))))))))))))).)...)))))))..))))... ( -35.95, z-score = -1.72, R) >droAna3.scaffold_12916 15717324 104 - 16180835 ---------------UCCUGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUGUCAGUUAAGCAUGAUAGCGAGUUGUGCGACCCGCCCCCA ---------------...((((((((((((((((((((.(((.((.....)).))).))).........((((....))))))))).))))))))))))......(((...)))..... ( -27.80, z-score = -2.03, R) >droPer1.super_49 177596 115 - 569410 ----ACGCGUCCGGUUCGGGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUAUGACAGCAAAUUGUGCGACCCGCCCUUG ----.............((((.(((..((..((((((((.....))..((((((((.((((((..............)))))).)))))))))))))).....))..)))..))))... ( -30.94, z-score = -1.25, R) >dp4.chr4_group4 4740605 115 + 6586962 ----ACGCGUCCGGUUCGGACUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUAUGACAGCAAAUUGUGCGACCCGCCCUUG ----..(((((((...))))).(((..((..((((((((.....))..((((((((.((((((..............)))))).)))))))))))))).....))..)))..))..... ( -30.44, z-score = -1.49, R) >droWil1.scaffold_180772 6409362 111 + 8906247 -----UCUUUCAAAUGC---CUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUGAAAUGGAAUCUAAUCACGUUCUAAUUUCUACUAAGUAUGACAGCAAAUUGUGUGAACAGUUGUCC -----............---(((((((((..(((((((....((((((.....((((((((((........))))).)))))...))))))))))))).....))))).))))...... ( -27.70, z-score = -2.03, R) >droVir3.scaffold_12723 3738000 94 - 5802038 ---------CCCCAAUUAAGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUUUGACAGCAAUUU---------------- ---------..........(((((((.((((((((..(((((.((.....)).....(((.......))).....))))).))))).))).))))))).....---------------- ( -20.70, z-score = -1.72, R) >droMoj3.scaffold_6500 9259341 112 + 32352404 ------AUACCCCAAUUAAGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUUUGACAGCAAUUUGUGUGCGACGACCAA- ------.............(.((.(((((..(((((((((((....))))((((((.((((((..............)))))).)))))).))))))).....))))))).)......- ( -21.64, z-score = 0.11, R) >droGri2.scaffold_15252 8081875 103 - 17193109 ---------CUCCAAUUAAGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUUUGACAGCAAAUUGUGUAAGCC------- ---------.........(((......)))((((..(((((.((((....((((((.((((((..............)))))).)))))).....)))))))))....))))------- ( -22.34, z-score = -1.13, R) >consensus _____CCGGCCAGCUUCCUGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUGUGACAGCAAAUUGUGCGACCCGCCCUCG ...................(((((((((((((((((((.(((.((.....)).))).))).....................))))).)))))))))))..................... (-21.27 = -21.59 + 0.32)
| Location | 15,919,187 – 15,919,303 |
|---|---|
| Length | 116 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.27 |
| Shannon entropy | 0.47407 |
| G+C content | 0.44274 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -17.85 |
| Energy contribution | -18.29 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15919187 116 - 23011544 ---GGGCGGGUCGCACAAUUCGCUGUCACACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCUAAACUGACAGCCAGCAUGACAGCAGGGAGCUUGCCGGGCAUG ---.((((((((.(.......(((((((....(((.(((((((.((......)).))))))).)))(((.....)))....)))))))..((......)).).)).))))))....... ( -30.60, z-score = -0.83, R) >droSim1.chr2L 15631988 119 - 22036055 CAAGGGCGGGUCGCACAAUUCGCUGUCACACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCGCAACUGACAGCCAGCAUGACAGCAGGGAGCUUCCCGGGCAUG ......((((..((....(..(((((((..(((((.(((((((.((......)).))))))).)))))......((((........))..)).)))))))..)..))..))))...... ( -30.70, z-score = -0.65, R) >droSec1.super_5 4653254 119 - 5866729 CAAGGGCGGGUCGCACAAUUCGCUGUCACACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCGCAACUGACAGCCAGCAUGACAGCAGGGAGCUUCCCCGGCAUG ....(.((((..((....(..(((((((..(((((.(((((((.((......)).))))))).)))))......((((........))..)).)))))))..)..))...)))).)... ( -31.10, z-score = -0.79, R) >droYak2.chr2L 2904908 119 + 22324452 UGGGGGGGGGGGUCACAAUUCGCUGUCACACUUAACCGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCGCAACUGACAGCCAGCAUGACAGCACAGAGCUUCCUGAGCAUG .....((((((.((.......(((((((.........((((((.((......)).))))))......((.....)).....)))))))..((......))...)).))))))....... ( -27.40, z-score = 0.16, R) >droEre2.scaffold_4929 7618290 118 - 26641161 CGAGGGCGGGUCACACGAUUCGCUGUCACACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCGCCACUGACAGCCAGCAUGACAGCA-GGAGCUGCCCGGGCAUG ...((((((.((.........(((((((...((((.(((((((.((......)).))))))).))))((.....)).....)))))))..((......)).-.)).))))))....... ( -34.40, z-score = -1.37, R) >droAna3.scaffold_12916 15717324 104 + 16180835 UGGGGGCGGGUCGCACAACUCGCUAUCAUGCUUAACUGACAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCAGGA--------------- .(((.(((...)))....)))(((.(((((((...(((...((((...((((.......))))....((.....))....)))))))..))))))).)))....--------------- ( -24.40, z-score = -0.74, R) >droPer1.super_49 177596 115 + 569410 CAAGGGCGGGUCGCACAAUUUGCUGUCAUACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCCCGAACCGGACGCGU---- ...((((..((((........(((((((...((((.(((((((.((......)).))))))).))))((.....)).....))))))).....)))).)))).............---- ( -28.62, z-score = -1.22, R) >dp4.chr4_group4 4740605 115 - 6586962 CAAGGGCGGGUCGCACAAUUUGCUGUCAUACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGUCCGAACCGGACGCGU---- .....((..((((........(((((((...((((.(((((((.((......)).))))))).))))((.....)).....))))))).....)))).(((((...)))))))..---- ( -27.82, z-score = -0.92, R) >droWil1.scaffold_180772 6409362 111 - 8906247 GGACAACUGUUCACACAAUUUGCUGUCAUACUUAGUAGAAAUUAGAACGUGAUUAGAUUCCAUUUCAGCAUUAAGCACAACUGACAGCCAGCAUGACAG---GCAUUUGAAAGA----- ((((....))))...(((..(((((((((.((((((.(((((..((((.......).))).)))))...)))))).....(((.....))).)))))).---))).))).....----- ( -19.20, z-score = 0.30, R) >droVir3.scaffold_12723 3738000 94 + 5802038 ----------------AAAUUGCUGUCAAACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCUUAAUUGGGG--------- ----------------.....(((((((..(((((.(((((((.((......)).))))))).)))))............(((.....)))..)))))))..........--------- ( -20.10, z-score = -1.48, R) >droMoj3.scaffold_6500 9259341 112 - 32352404 -UUGGUCGUCGCACACAAAUUGCUGUCAAACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCUUAAUUGGGGUAU------ -...(((((.((.........(((((((...((((.(((((((.((......)).))))))).))))((.....)).....)))))))..))))))).((((.....))))..------ ( -22.10, z-score = -0.04, R) >droGri2.scaffold_15252 8081875 103 + 17193109 -------GGCUUACACAAUUUGCUGUCAAACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCUUAAUUGGAG--------- -------........(((((.(((((((..(((((.(((((((.((......)).))))))).)))))............(((.....)))..)))))))..)))))...--------- ( -23.70, z-score = -2.20, R) >consensus CAAGGGCGGGUCGCACAAUUCGCUGUCACACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCAGGAAGCUGCCCGG_____ .....................(((((((..(((((.(((((((.((......)).))))))).)))))............(((.....)))..)))))))................... (-17.85 = -18.29 + 0.44)
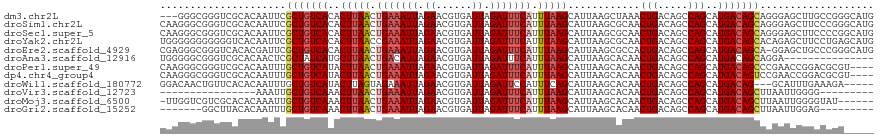
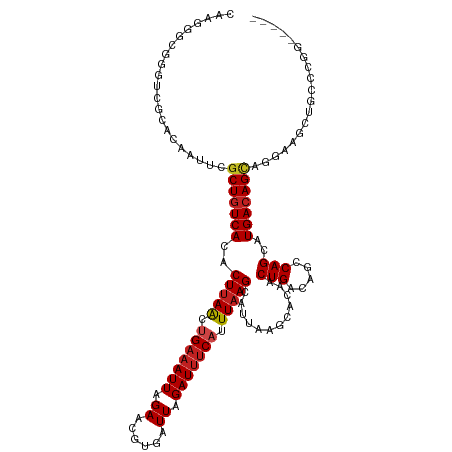
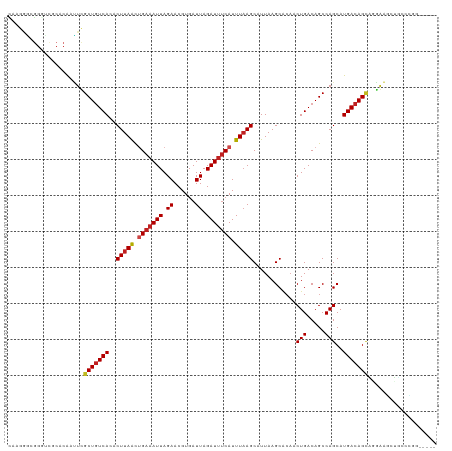
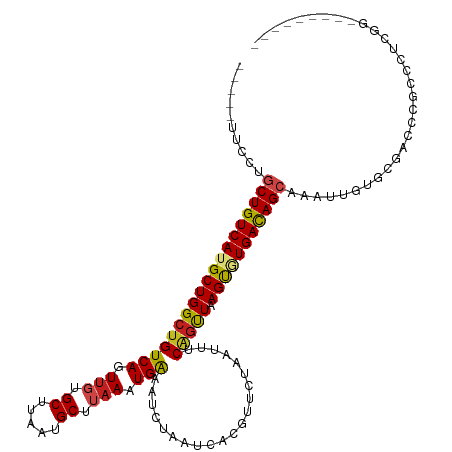
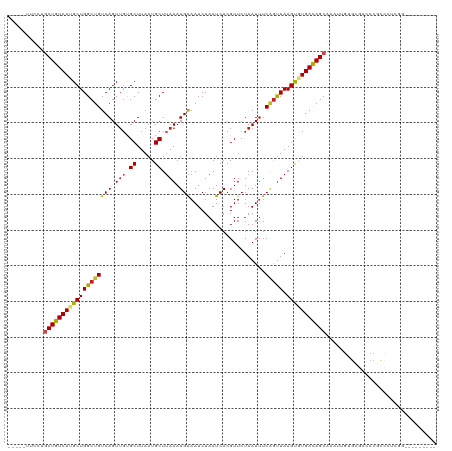
| Location | 15,919,196 – 15,919,305 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.28 |
| Shannon entropy | 0.45068 |
| G+C content | 0.42889 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.59 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15919196 109 + 23011544 CAAGCUCCCUGCUGUCAUGCUGGCUGUCAGUUUAGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUGUGACAGCGAAUUGUGCGACCCGCCCUC----------- ...((.(..(((((((((...(((((......)))))....((((((.((((((..............)))))).)))))))))))))))....).))...........----------- ( -26.04, z-score = -1.44, R) >droSim1.chr2L 15631997 120 + 22036055 GAAGCUCCCUGCUGUCAUGCUGGCUGUCAGUUGCGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUGUGACAGCGAAUUGUGCGACCCGCCCUUGGAAUACGUUC (((((.....))).))..((.((.((((((((.((((..((((((((.((((((..............)))))).))))))))...))))))))).))).)).))............... ( -27.44, z-score = -0.21, R) >droSec1.super_5 4653263 120 + 5866729 GAAGCUCCCUGCUGUCAUGCUGGCUGUCAGUUGCGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUGUGACAGCGAAUUGUGCGACCCGCCCUUGGAAUACGCUC ..((((((.(((((((((((((((((..(((((.((.....)).....(((.......))).....))))).))))).))))))))))))......(((...)))....)))....))). ( -27.70, z-score = -0.19, R) >droYak2.chr2L 2904917 120 - 22324452 GAAGCUCUGUGCUGUCAUGCUGGCUGUCAGUUGCGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCGGUUAAGUGUGACAGCGAAUUGUGACCCCCCCCCCCAAACCCCCCUG ...((....(((((((((((((((((..(((((.((.....)).....(((.......))).....))))).))))).))))))))))))....))........................ ( -24.30, z-score = -1.45, R) >droEre2.scaffold_4929 7618299 119 + 26641161 GCAGCUCC-UGCUGUCAUGCUGGCUGUCAGUGGCGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUGUGACAGCGAAUCGUGUGACCCGCCCUCGGAAUACCCUC ((((...)-))).((((((((.((((((....(((((((((.......((((((..............))))))))))))))))))))).)...)))))))(((....)))......... ( -29.75, z-score = -0.77, R) >droAna3.scaffold_12916 15717324 114 - 16180835 ------UCCUGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUGUCAGUUAAGCAUGAUAGCGAGUUGUGCGACCCGCCCCCAGAACACGAUU ------...((((((((((((((((((((.(((.((.....)).))).))).........((((....))))))))).))))))))))))(((((((................))))))) ( -29.89, z-score = -1.60, R) >droPer1.super_49 177606 106 - 569410 -----UUCGGGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUAUGACAGCAAAUUGUGCGACCCGCCCUUGG--------- -----...((((.(((..((..((((((((.....))..((((((((.((((((..............)))))).)))))))))))))).....))..)))..))))....--------- ( -30.94, z-score = -2.44, R) >dp4.chr4_group4 4740615 106 + 6586962 -----UUCGGACUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUAUGACAGCAAAUUGUGCGACCCGCCCUUGG--------- -----...((...(((..((..((((((((.....))..((((((((.((((((..............)))))).)))))))))))))).....))..)))...)).....--------- ( -25.14, z-score = -0.95, R) >droWil1.scaffold_180772 6409371 103 + 8906247 --------UGCCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUGAAAUGGAAUCUAAUCACGUUCUAAUUUCUACUAAGUAUGACAGCAAAUUGUGUGAACAGUUGUCCA--------- --------...(((((((((..(((((((....((((((.....((((((((((........))))).)))))...))))))))))))).....))))).)))).......--------- ( -27.70, z-score = -1.93, R) >droVir3.scaffold_12723 3738006 88 - 5802038 ------UUAAGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUUUGACAGCAAUUU-------------------------- ------....(((((((.((((((((..(((((.((.....)).....(((.......))).....))))).))))).))).))))))).....-------------------------- ( -20.70, z-score = -1.64, R) >droMoj3.scaffold_6500 9259350 103 + 32352404 ------UUAAGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUUUGACAGCAAUUUGUGUGCGACGACCAA----------- ------....(.((.(((((..(((((((((((....))))((((((.((((((..............)))))).)))))).))))))).....))))))).)......----------- ( -21.64, z-score = 0.17, R) >droGri2.scaffold_15252 8081881 97 - 17193109 ------UUAAGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUUUGACAGCAAAUUGUGUAAGCC----------------- ------...(((......)))((((..(((((.((((....((((((.((((((..............)))))).)))))).....)))))))))....))))----------------- ( -22.34, z-score = -1.24, R) >consensus _____UUCCUGCUGUCAUGCUGGCUGUCAGUUGUGCUUAAUGCUUAAAUGAAAUCUAAUCACGUUCUAAUUUCAGUUAAGUGUGACAGCAAAUUGUGCGACCCGCCCUCGG_________ ..........(((((((((((((((((((.(((.((.....)).))).))).....................))))).)))))))))))............................... (-21.27 = -21.59 + 0.32)
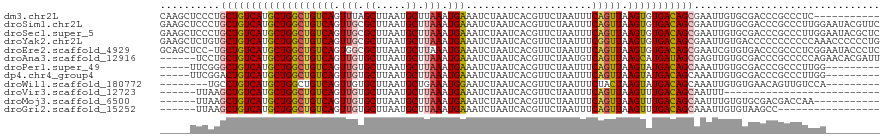
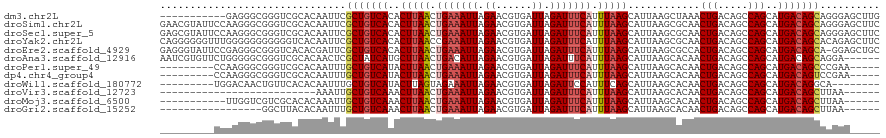
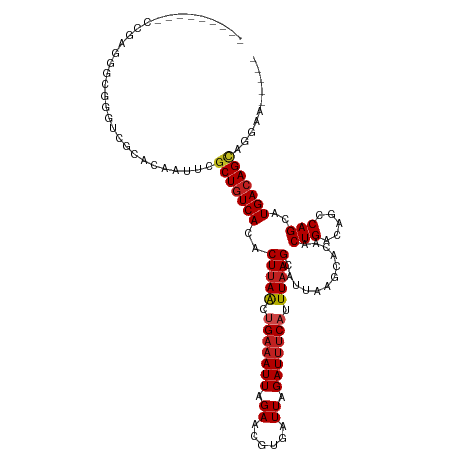
| Location | 15,919,196 – 15,919,305 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.28 |
| Shannon entropy | 0.45068 |
| G+C content | 0.42889 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -17.85 |
| Energy contribution | -18.29 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15919196 109 - 23011544 -----------GAGGGCGGGUCGCACAAUUCGCUGUCACACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCUAAACUGACAGCCAGCAUGACAGCAGGGAGCUUG -----------((.(((((((......))))))).))...(((((.(((((((.((......)).))))))).)))))...((((((...(((....(......)....)))..)))))) ( -26.10, z-score = -0.70, R) >droSim1.chr2L 15631997 120 - 22036055 GAACGUAUUCCAAGGGCGGGUCGCACAAUUCGCUGUCACACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCGCAACUGACAGCCAGCAUGACAGCAGGGAGCUUC (((.((.((((....(((...))).......(((((((..(((((.(((((((.((......)).))))))).)))))......((((........))..)).))))))).))))))))) ( -27.50, z-score = -0.31, R) >droSec1.super_5 4653263 120 - 5866729 GAGCGUAUUCCAAGGGCGGGUCGCACAAUUCGCUGUCACACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCGCAACUGACAGCCAGCAUGACAGCAGGGAGCUUC ((((...((((....(((...))).......(((((((..(((((.(((((((.((......)).))))))).)))))......((((........))..)).))))))).)))))))). ( -29.30, z-score = -0.51, R) >droYak2.chr2L 2904917 120 + 22324452 CAGGGGGGUUUGGGGGGGGGGGUCACAAUUCGCUGUCACACUUAACCGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCGCAACUGACAGCCAGCAUGACAGCACAGAGCUUC ...((((.((((.(((..(......)..)))(((((((..(((((..((((((.((......)).))))))..)))))......((((........))..)).))))))).)))).)))) ( -26.10, z-score = 0.12, R) >droEre2.scaffold_4929 7618299 119 - 26641161 GAGGGUAUUCCGAGGGCGGGUCACACGAUUCGCUGUCACACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCGCCACUGACAGCCAGCAUGACAGCA-GGAGCUGC .(((((.....((.((((((((....)))))))).))...(((((.(((((((.((......)).))))))).)))))........))).))..(((((.((......)).-.).)))). ( -31.80, z-score = -0.98, R) >droAna3.scaffold_12916 15717324 114 + 16180835 AAUCGUGUUCUGGGGGCGGGUCGCACAACUCGCUAUCAUGCUUAACUGACAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCAGGA------ ..(((((..((((.(((((((......)))))))...((((((((.(((.(((.((......)).))).))).))))))))................)))))))))........------ ( -28.00, z-score = -0.47, R) >droPer1.super_49 177606 106 + 569410 ---------CCAAGGGCGGGUCGCACAAUUUGCUGUCAUACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCCCGAA----- ---------....((((..((((........(((((((...((((.(((((((.((......)).))))))).))))((.....)).....))))))).....)))).))))...----- ( -28.62, z-score = -2.44, R) >dp4.chr4_group4 4740615 106 - 6586962 ---------CCAAGGGCGGGUCGCACAAUUUGCUGUCAUACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGUCCGAA----- ---------....((((..((((........(((((((...((((.(((((((.((......)).))))))).))))((.....)).....))))))).....)))).))))...----- ( -25.92, z-score = -1.55, R) >droWil1.scaffold_180772 6409371 103 - 8906247 ---------UGGACAACUGUUCACACAAUUUGCUGUCAUACUUAGUAGAAAUUAGAACGUGAUUAGAUUCCAUUUCAGCAUUAAGCACAACUGACAGCCAGCAUGACAGGCA-------- ---------(((((....)))))........(((((((.........(((((..((((.......).))).))))).((.....)).....)))))))..((.......)).-------- ( -17.90, z-score = 0.43, R) >droVir3.scaffold_12723 3738006 88 + 5802038 --------------------------AAAUUGCUGUCAAACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCUUAA------ --------------------------.....(((((((..(((((.(((((((.((......)).))))))).)))))............(((.....)))..)))))))....------ ( -20.10, z-score = -2.38, R) >droMoj3.scaffold_6500 9259350 103 - 32352404 -----------UUGGUCGUCGCACACAAAUUGCUGUCAAACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCUUAA------ -----------...(((((.((.........(((((((...((((.(((((((.((......)).))))))).))))((.....)).....)))))))..))))))).......------ ( -21.50, z-score = -0.83, R) >droGri2.scaffold_15252 8081881 97 + 17193109 -----------------GGCUUACACAAUUUGCUGUCAAACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCUUAA------ -----------------..............(((((((..(((((.(((((((.((......)).))))))).)))))............(((.....)))..)))))))....------ ( -20.10, z-score = -1.62, R) >consensus _________CCGAGGGCGGGUCGCACAAUUCGCUGUCACACUUAACUGAAAUUAGAACGUGAUUAGAUUUCAUUUAAGCAUUAAGCACAACUGACAGCCAGCAUGACAGCAGGAA_____ ...............................(((((((..(((((.(((((((.((......)).))))))).)))))............(((.....)))..))))))).......... (-17.85 = -18.29 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:51 2011