
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,876,287 – 15,876,380 |
| Length | 93 |
| Max. P | 0.715833 |

| Location | 15,876,287 – 15,876,380 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 74.35 |
| Shannon entropy | 0.52055 |
| G+C content | 0.33323 |
| Mean single sequence MFE | -21.68 |
| Consensus MFE | -6.87 |
| Energy contribution | -7.15 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
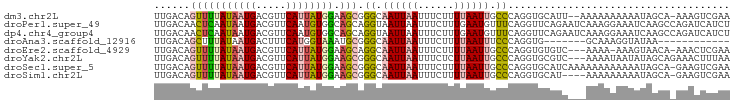
>dm3.chr2L 15876287 93 - 23011544 UUGACAGUUUUAUAAUGACGUUCAUUAUGGAAGCGGGCAAUUAAUUUCUUUUAAUUGCCCAGGUGCAUU--AAAAAAAAAAUAGCA-AAAGUCGAA (((((.(((((((((((.....)))))).)))))((((((((((......))))))))))...((((((--........))).)))-...))))). ( -28.00, z-score = -4.78, R) >droPer1.super_49 129489 96 + 569410 UUGACAACUCAAUAAUGACGUUCAAUGUGGCAGCAGGUAAUUAAUUUCUUUGAAUGUUUCAGGUUCAGAAUCAAAGGAAAUCAAGCCAGAUCAUCU .(((..(((((....))).)))))((.((((....(((......(((((((((....(((.......)))))))))))))))..)))).))..... ( -12.00, z-score = 1.78, R) >dp4.chr4_group4 4694515 96 - 6586962 UUGACAACUCAAUAAUGACGUUCAAUGUGGCAGCAGGUAAUUAAUUUCUUUGAAUGUUUCAGGUUCAGAAUCAAAGGAAAUCAAGCCAGAUCAUCU .(((..(((((....))).)))))((.((((....(((......(((((((((....(((.......)))))))))))))))..)))).))..... ( -12.00, z-score = 1.78, R) >droAna3.scaffold_12916 15676158 77 + 16180835 UUGACAGCUUUAUAAUGACUUUCAUGGUAAAUGCGGGCAAUUAAUUUCUUUUAAUUUCCCAGGUG-------GCAAAGGUAUAA------------ (((.(((((((((.(((.....))).))))).))(((.((((((......)))))).)))...))-------.)))........------------ ( -15.30, z-score = -0.73, R) >droEre2.scaffold_4929 7574731 91 - 26641161 UUGACAGUUUUAUAAUGACGUUCAUUAUGGAAGCAGGCAAUUAAUUUCUUUUAAUUGCCCAGGUGUGUC---AAAA-AAAGUAACA-AAACUCGAA ((((((.((((((((((.....))))))))))((.(((((((((......)))))))))...)).))))---))..-.........-......... ( -24.10, z-score = -3.55, R) >droYak2.chr2L 2854254 93 + 22324452 UUGACAGUUUUAUAAUGACGUUCAUUAUGGAAGCGGGCAAUUAAUUUCUCUUAAUUGCCCAGGUGCGUC---AAAAUAAUAUAGCAGAAACUUUAA (((((.(((((((((((.....)))))).)))))((((((((((......))))))))))......)))---))...................... ( -26.80, z-score = -4.04, R) >droSec1.super_5 4607609 95 - 5866729 UUGACAGUUUUAUAAUGACGUUCAUUAUGGAAGCGGGCAAUUAAUUUCUUUUAAUUGCCCAGGUGCAUCAAAAAAAAAAAAUAGCA-GAAGUCGAA (((((..((((((((((.....))))))))))((((((((((((......)))))))))).((....))..............)).-...))))). ( -27.70, z-score = -4.43, R) >droSim1.chr2L 15585205 91 - 22036055 UUGACAGUUUUAUAAUGACGUUCAUUAUGGAAGCGGGCAAUUAAUUUCUUUUAAUUGCCCAGGUGCAU----AAAAAAAAAUAGCA-GAAGUCGAA (((((.(((((((((((.....)))))).)))))((((((((((......))))))))))...(((..----...........)))-...))))). ( -27.52, z-score = -4.60, R) >consensus UUGACAGUUUUAUAAUGACGUUCAUUAUGGAAGCGGGCAAUUAAUUUCUUUUAAUUGCCCAGGUGCAUC___AAAAAAAAAUAACA_AAAGUCGAA ......(((((((((((.....)))))))).))).((.((((((......)))))).))..................................... ( -6.87 = -7.15 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:46 2011