| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,872,291 – 15,872,396 |
| Length | 105 |
| Max. P | 0.895180 |

| Location | 15,872,291 – 15,872,396 |
|---|---|
| Length | 105 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Shannon entropy | 0.34487 |
| G+C content | 0.41447 |
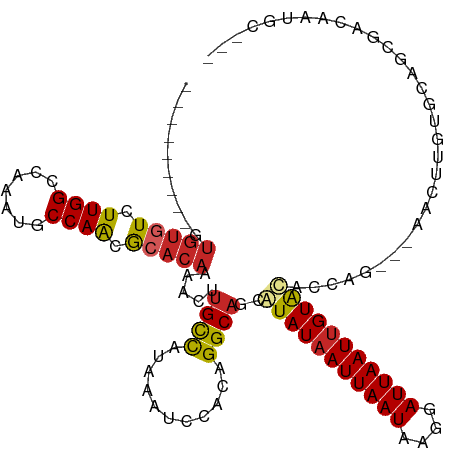
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -18.24 |
| Energy contribution | -18.28 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895180 |
| Prediction | RNA |
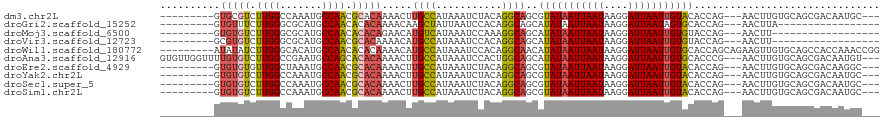
Download alignment: ClustalW | MAF
>dm3.chr2L 15872291 105 + 23011544 ---------GUGCGUCUUGGCCAAAUGCCAACGCACAAAACUUGCCAUAAAUCUACAGGCAGCGUAUAAUUAAUAAGGAUUAAUUGUACACCAG---AACUUGUGCAGCGACAAUGC--- ---------....((((((((.....))))).((((((...(((((...........))))).(((((((((((....))))))))))).....---...))))))...))).....--- ( -30.50, z-score = -2.54, R) >droGri2.scaffold_15252 8034094 91 - 17193109 ---------GUGUUUCUUGGGCGCAUGCCAACACACAAAACAAGCUAUUAAUCCACAGGCAGCAUAUAAUUAAUAAGGAUUAAUAGUGCACCAG---AACUUA----------------- ---------((((......(((....)))...)))).......(((((((((((......................))))))))))).......---......----------------- ( -18.45, z-score = -1.07, R) >droMoj3.scaffold_6500 9199992 90 + 32352404 ---------GUGUGUCUUGGGCGCAUGCCAACACACAGAACAUGUCAUAAAUCCAAAGGCAGCAUAUAAUUAAUAAGGAUUAAUUGUGUACCAG---AACUU------------------ ---------((((((....(((....))).))))))......((((...........))))..(((((((((((....))))))))))).....---.....------------------ ( -19.90, z-score = -0.79, R) >droVir3.scaffold_12723 3665424 90 - 5802038 ---------GCGUGUCUUGGGCGCAUGCCAACGCACAAAACAUGCCAUAAAUCCACAGGCAGCAUAUAAUUAAUAAGGAUUAAUUGUGUACCAG---AACUU------------------ ---------((((((.((((.......)))).))))......((((...........))))))(((((((((((....))))))))))).....---.....------------------ ( -22.10, z-score = -1.11, R) >droWil1.scaffold_180772 6336337 111 + 8906247 ---------AUAUAUCUUGGGCACAUGCCAACACACAAAACAUGCCAUAAAUCCACAGGCAACAUAUAAUUAAUAAGGAUUAAUUGUGCACCAGCAGAAGUUGUGCAGCCACCAAACCGG ---------.......((((.....(((((((..........((((...........))))...((((((((((....))))))))))...........)))).)))....))))..... ( -22.00, z-score = -0.23, R) >droAna3.scaffold_12916 15673053 114 - 16180835 GUGUUGGUUUUGUGUCUUGGCCGAAUGCCAGCACACAAAACUUGCCAUAAAUCCACUGGCAGCAUAUAAUUAAUAAGGAUUAAUUGUGCACCCG---AACUUGUGCAGCGACAAUGU--- .((((((((((((((.(((((.....))))).))))))))))(((((.........)))))((..(((((((((....)))))))))((((...---.....)))).))))))....--- ( -36.40, z-score = -2.42, R) >droEre2.scaffold_4929 7570728 105 + 26641161 ---------GUGUGUGUUGGCUAAAUGCCAACGCACAAAACUUGCCAUAAAUCUACAGGCAGCGUAUAAUUAAUAAGGAUUAAUUGUACACCAG---AACUUGUGCAGCGACAAGGC--- ---------.(((((((((((.....))))))))))).....((((...........))))(((((((((((((....))))))))))).....---..(((((......)))))))--- ( -37.50, z-score = -4.29, R) >droYak2.chr2L 2850153 105 - 22324452 ---------GUGUGUCUUGGCCAAAUGCCAACGCACAAAACUUGCCAUAAAUCUACAGGCAGCGUAUAAUUAAUAAGGAUUAAUUGUACACCAG---AACUUGUGCAGCGACAAUGC--- ---------...(((((((((.....))))).((((((...(((((...........))))).(((((((((((....))))))))))).....---...))))))...))))....--- ( -31.10, z-score = -2.64, R) >droSec1.super_5 4603746 105 + 5866729 ---------GUGUGUCUUGGCCAAAUGCCAACGCACAAAACUUGCCAUAAAUCUACAGGCAGCGUAUAAUUAAUAAGGAUUAAUUGUACACCAG---AACUUGUGCAGCGACAAUGC--- ---------...(((((((((.....))))).((((((...(((((...........))))).(((((((((((....))))))))))).....---...))))))...))))....--- ( -31.10, z-score = -2.64, R) >droSim1.chr2L 15581238 105 + 22036055 ---------GUGUGUCUUGGCCAAAUGCCAACGCACAAAACUUGCCAUAAAUCUACAGGCAGCGUAUAAUUAAUAAGGAUUAAUUGUACACCAG---AACUUGUGCAGCGACAAUGC--- ---------...(((((((((.....))))).((((((...(((((...........))))).(((((((((((....))))))))))).....---...))))))...))))....--- ( -31.10, z-score = -2.64, R) >consensus _________GUGUGUCUUGGCCAAAUGCCAACGCACAAAACUUGCCAUAAAUCCACAGGCAGCAUAUAAUUAAUAAGGAUUAAUUGUACACCAG___AACUUGUGCAGCGACAAUGC___ ..........(((((.((((.......)))).))))).....((((...........))))..(((((((((((....)))))))))))............................... (-18.24 = -18.28 + 0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:45 2011