| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,871,649 – 15,871,741 |
| Length | 92 |
| Max. P | 0.841361 |

| Location | 15,871,649 – 15,871,741 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.13 |
| Shannon entropy | 0.38177 |
| G+C content | 0.41757 |
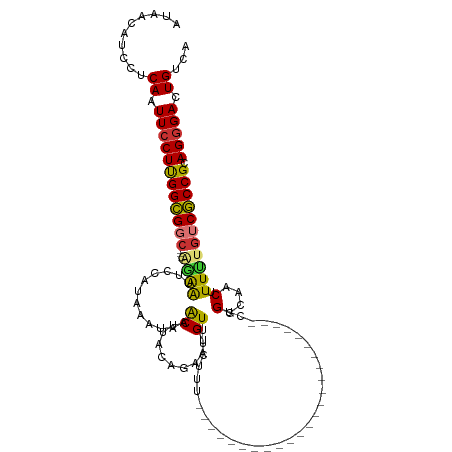
| Mean single sequence MFE | -24.96 |
| Consensus MFE | -11.58 |
| Energy contribution | -11.64 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
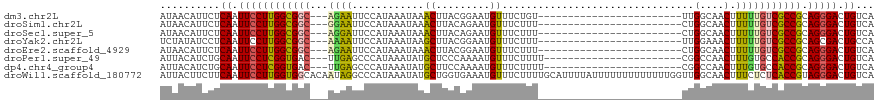
>dm3.chr2L 15871649 92 + 23011544 UGACAGUCCCUGCGGCGACAAAAAGUUGCCAA------------------------ACAGAAACAUUCCGUAAGUUUAUUUAUGGAAUUCU---GCCGCCAAGGAAUUGAGAAUGUUAU ...((((.(((((((((((.....))))))..------------------------.((((...((((((((((....)))))))))))))---)..))..))).)))).......... ( -28.30, z-score = -3.87, R) >droSim1.chr2L 15580604 92 + 22036055 UGACAGUCCCUGCGGCGACAAAAAGUUGCCAG------------------------AAAGAAACAUUCUGUAAGUUUAUUUAUGGAAUUCC---GCCGCCAAGGAAUUGAGAAUGUUAU ...((((.(((((((((.......(((.....------------------------.....)))((((((((((....))))))))))..)---)))))..))).)))).......... ( -23.30, z-score = -2.21, R) >droSec1.super_5 4603108 92 + 5866729 UGACAGUCCCUGCGGCGACAAAAAGUUGCCAG------------------------AAAGAAACAUUCUGUAAGUUUAUUUAUGGAAUCCU---GCCGCCAAGGAAUUGAGAAUGUUAU ...((((.(((((((((((.....))))))..------------------------........((((((((((....))))))))))...---...))..))).)))).......... ( -22.00, z-score = -1.74, R) >droYak2.chr2L 2849511 92 - 22324452 UGGCAGUCGCUGCGGCGACAAAAAGUUUCCAA------------------------AAAGAAACAUUCCGUAAGCUUAUUUAUGGAAUUUU---GCCGCCAAGGAAUUGAGGAUAUAGA ((((.(((((....))))).....(((((...------------------------...)))))((((((((((....))))))))))...---...)))).................. ( -26.20, z-score = -2.24, R) >droEre2.scaffold_4929 7570082 92 + 26641161 UGACAGUCCCUGCGGCGACAAAAAGUUGCCAG------------------------AAAGAAACAUUCCGUAAGUUUAUUUAUGGAAUUCU---GCCGCCAAGGAAUUGAGAAUGUUAU ...((((.(((((((((((.....)))))).(------------------------..(((...((((((((((....)))))))))))))---..)))..))).)))).......... ( -26.40, z-score = -3.29, R) >droPer1.super_49 124062 93 - 569410 UGACAGUCCCUGCGGUGGCACAAAGUUGGCCG-----------------------AAAAGAAACAUUUUGGGAGCAUAUUUAUGGGCUCAA---GUCACCGAGGAAUUGCAGAUGUAAU ((.((((.(((.(((((((..........(((-----------------------(((.......))))))((((..........))))..---)))))))))).))))))........ ( -28.50, z-score = -1.99, R) >dp4.chr4_group4 4689045 93 + 6586962 UGACAGUCCCUGCGGUGGCACAAAGUUGGCCG-----------------------AAAAGAAACAUUUUGGAAGCAUAUUUAUGGGCUCAA---GUCACCGAGGAAUUGCAGAUGUAAU ((.((((.(((.(((((((.....(((..(((-----------------------(((.......)))))).))).......((....)).---)))))))))).))))))........ ( -24.60, z-score = -0.97, R) >droWil1.scaffold_180772 6335764 119 + 8906247 UGACAGUCCCUACGGUGAGAGAAAGUUGCCAACCAAAAAAAAAAAAAUAAAAUGCAAAAGAAACAUUUCACCAGCAUAUUUAUGGGCCUAUUGUGCCACCAAGGAAUUGAAGAAGUAAU ...((((.(((..(((((((.....((((........................))))........)))))))............((((....).)))....))).)))).......... ( -20.38, z-score = -0.84, R) >consensus UGACAGUCCCUGCGGCGACAAAAAGUUGCCAG________________________AAAGAAACAUUCCGUAAGCUUAUUUAUGGAAUUCU___GCCGCCAAGGAAUUGAGAAUGUAAU ...((((.(((..((((((.....))))))....................................................(((.............)))))).)))).......... (-11.58 = -11.64 + 0.06)



| Location | 15,871,649 – 15,871,741 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.13 |
| Shannon entropy | 0.38177 |
| G+C content | 0.41757 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -12.52 |
| Energy contribution | -12.11 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.841361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15871649 92 - 23011544 AUAACAUUCUCAAUUCCUUGGCGGC---AGAAUUCCAUAAAUAAACUUACGGAAUGUUUCUGU------------------------UUGGCAACUUUUUGUCGCCGCAGGGACUGUCA ..........((.((((((((((((---((((((((.(((......))).)))))........------------------------..(....)..)))))))))).))))).))... ( -26.10, z-score = -2.87, R) >droSim1.chr2L 15580604 92 - 22036055 AUAACAUUCUCAAUUCCUUGGCGGC---GGAAUUCCAUAAAUAAACUUACAGAAUGUUUCUUU------------------------CUGGCAACUUUUUGUCGCCGCAGGGACUGUCA ..........((.((((((((((((---((((..(((.(((.((((.........)))).)))------------------------.))).....))))))))))).))))).))... ( -25.10, z-score = -2.98, R) >droSec1.super_5 4603108 92 - 5866729 AUAACAUUCUCAAUUCCUUGGCGGC---AGGAUUCCAUAAAUAAACUUACAGAAUGUUUCUUU------------------------CUGGCAACUUUUUGUCGCCGCAGGGACUGUCA ..........((.((((((((((((---((((..(((.(((.((((.........)))).)))------------------------.))).....))))))))))).))))).))... ( -25.50, z-score = -3.02, R) >droYak2.chr2L 2849511 92 + 22324452 UCUAUAUCCUCAAUUCCUUGGCGGC---AAAAUUCCAUAAAUAAGCUUACGGAAUGUUUCUUU------------------------UUGGAAACUUUUUGUCGCCGCAGCGACUGCCA ...................((((((---((((((((.(((......))).)))))((((((..------------------------..))))))..)))))))))((((...)))).. ( -26.90, z-score = -3.58, R) >droEre2.scaffold_4929 7570082 92 - 26641161 AUAACAUUCUCAAUUCCUUGGCGGC---AGAAUUCCAUAAAUAAACUUACGGAAUGUUUCUUU------------------------CUGGCAACUUUUUGUCGCCGCAGGGACUGUCA ..........((.((((((((((((---((((((((.(((......))).)))))........------------------------..(....)..)))))))))).))))).))... ( -26.10, z-score = -3.34, R) >droPer1.super_49 124062 93 + 569410 AUUACAUCUGCAAUUCCUCGGUGAC---UUGAGCCCAUAAAUAUGCUCCCAAAAUGUUUCUUUU-----------------------CGGCCAACUUUGUGCCACCGCAGGGACUGUCA .........(((.(((((((((...---..((((..........))))................-----------------------.(((((....)).))))))).))))).))).. ( -21.30, z-score = -1.07, R) >dp4.chr4_group4 4689045 93 - 6586962 AUUACAUCUGCAAUUCCUCGGUGAC---UUGAGCCCAUAAAUAUGCUUCCAAAAUGUUUCUUUU-----------------------CGGCCAACUUUGUGCCACCGCAGGGACUGUCA .........(((.((((((((((((---(((.(((...((((((.........)))))).....-----------------------.))))))....))..))))).))))).))).. ( -19.20, z-score = -0.32, R) >droWil1.scaffold_180772 6335764 119 - 8906247 AUUACUUCUUCAAUUCCUUGGUGGCACAAUAGGCCCAUAAAUAUGCUGGUGAAAUGUUUCUUUUGCAUUUUAUUUUUUUUUUUUUGGUUGGCAACUUUCUCUCACCGUAGGGACUGUCA ..........((.(((((((((((........(((..((((......((((((((((.......))))))))))........))))...))).........)))))).))))).))... ( -24.47, z-score = -0.86, R) >consensus AUAACAUCCUCAAUUCCUUGGCGGC___AGAAUUCCAUAAAUAAACUUACAGAAUGUUUCUUU________________________CUGGCAACUUUUUGUCGCCGCAGGGACUGUCA ..........((.((((((((((((...................((.........))................................(....).....))))))).))))).))... (-12.52 = -12.11 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:44 2011