| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,818,218 – 15,818,312 |
| Length | 94 |
| Max. P | 0.968066 |

| Location | 15,818,218 – 15,818,312 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 67.85 |
| Shannon entropy | 0.64311 |
| G+C content | 0.45444 |
| Mean single sequence MFE | -24.99 |
| Consensus MFE | -9.49 |
| Energy contribution | -11.13 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
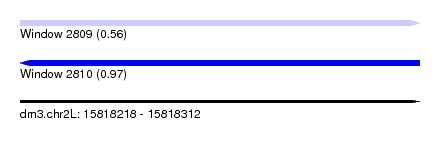
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.564389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
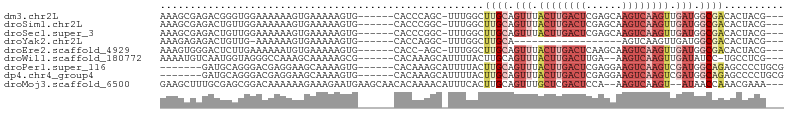
>dm3.chr2L 15818218 94 + 23011544 AAAGCGAGACGGGUGGAAAAAAGUGAAAAAGUG------CACCCAGC-UUUGGCUUGCAGUUUACUUGACUCGAGCAAGUCAAGUUGAUGGCGACACUACG--- .((((.(((((((((.(......(....)..).------))))).).-))).))))((.(((.((((((((......)))))))).))).)).........--- ( -26.30, z-score = -0.79, R) >droSim1.chr2L 15527721 94 + 22036055 AAAGCGAGACUGUUGGAAAAAAGUGAAAAAGUG------CACCCGGC-UUUGGCUUGCAGUUUACUUGACUCGAGCAAGUCAAGUUGAUGGCGACACUACG--- ........(((.((....)).))).....((((------(.((((((-((.(((((((((((.....))))...)))))))))))))..)).).))))...--- ( -24.10, z-score = -0.14, R) >droSec1.super_3 2026938 94 + 7220098 AAAGCGAGACUGUUGGAAAAAAGUGAAAAAGUG------CACCCGGC-UUUGGCUUGCAGUUUACUUGACUCGAGCAAGUCAAGUUGAUGGCGACACUACG--- ........(((.((....)).))).....((((------(.((((((-((.(((((((((((.....))))...)))))))))))))..)).).))))...--- ( -24.10, z-score = -0.14, R) >droYak2.chr2L 2790687 75 - 22324452 AAAGAGAGACUGUUG-AAAAAAGUGAAAAAGUG------CACCAGGC-UUUGGCUUGCA------------------AGUCAAGUUGAUGGCGACACUACG--- ........(((.((.-...))))).....((((------(.(((.((-((.((((....------------------))))))))...))).).))))...--- ( -14.10, z-score = 0.68, R) >droEre2.scaffold_4929 7514421 93 + 26641161 AAAGUGGGACUCUUGAAAAAAUGUGAAAAAGUG------CACC-AGC-UUUGGCUUGCAGUUUACUUGACUCAAGCAAGUCAAGUUGAUGGCGACACUACG--- ...((((.((..((....))..))......(((------(..(-(((-((.(((((((((((.....))))...)))))))))))))...)).)).)))).--- ( -25.10, z-score = -1.01, R) >droWil1.scaffold_180772 2641236 92 - 8906247 AAAAUGUCAAUGGUAGGGCCAAAGCAAAAAGCG------CACAAAGCAUUUUACUUGCAGUUUACUUGACUUGA--AAGUCAAGUUGAUAUCC-UGCCUCG--- ...........(((((((.....((((((((.(------(.....)).))))..)))).(((.((((((((...--.)))))))).))).)))-))))...--- ( -25.90, z-score = -2.45, R) >droPer1.super_116 87803 91 + 110639 -------GAUGCAGGGACGAGGAAGCAAAAGUG------CACAAAGCAUUUUACUUGCAGUUUACUUGACUCGAGGAAGUCAAGUCGAUGGCAGAGCCCCUGCG -------...((((((.(.........((((((------(.....)))))))..((((.(((.((((((((......)))))))).))).)))).).)))))). ( -33.70, z-score = -3.40, R) >dp4.chr4_group4 4627594 91 + 6586962 -------GAUGCAGGGACGAGGAAGCAAAAGUG------CACAAAGCAUUUUACUUGCAGUUUACUUGACUCGAGGAAGUCAAGUCGAUGGCAGAGCCCCUGCG -------...((((((.(.........((((((------(.....)))))))..((((.(((.((((((((......)))))))).))).)))).).)))))). ( -33.70, z-score = -3.40, R) >droMoj3.scaffold_6500 23228466 97 - 32352404 GAAGCUUUGCGAGCGGACAAAAAAGAAAGAAUGAAGCAACACAAAACAUUUCACUUGCAGUUUGCUCGACUCCA--AAGUCAAGU--AUAACCAAACGAAA--- ...(((((((((((((((....(((...(((((.............)))))..)))...)))))))))....))--)))).....--..............--- ( -17.92, z-score = -0.82, R) >consensus AAAGCGAGACUGUUGGAAAAAAGUGAAAAAGUG______CACCAAGC_UUUGGCUUGCAGUUUACUUGACUCGAGCAAGUCAAGUUGAUGGCGACACUACG___ ......................................................((((.(((.((((((((......)))))))).))).)))).......... ( -9.49 = -11.13 + 1.64)

| Location | 15,818,218 – 15,818,312 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 67.85 |
| Shannon entropy | 0.64311 |
| G+C content | 0.45444 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -9.24 |
| Energy contribution | -9.92 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.968066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
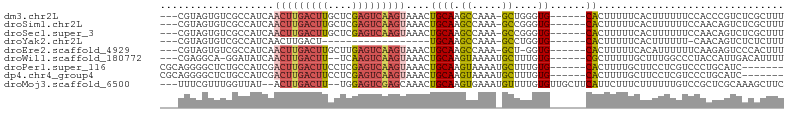
>dm3.chr2L 15818218 94 - 23011544 ---CGUAGUGUCGCCAUCAACUUGACUUGCUCGAGUCAAGUAAACUGCAAGCCAAA-GCUGGGUG------CACUUUUUCACUUUUUUCCACCCGUCUCGCUUU ---.(((((..........(((((((((....)))))))))..)))))((((...(-(.((((((------..................)))))).)).)))). ( -24.47, z-score = -1.85, R) >droSim1.chr2L 15527721 94 - 22036055 ---CGUAGUGUCGCCAUCAACUUGACUUGCUCGAGUCAAGUAAACUGCAAGCCAAA-GCCGGGUG------CACUUUUUCACUUUUUUCCAACAGUCUCGCUUU ---...((((.((((....(((((((((....)))))))))...(.((........-)).)))))------))))............................. ( -22.20, z-score = -1.06, R) >droSec1.super_3 2026938 94 - 7220098 ---CGUAGUGUCGCCAUCAACUUGACUUGCUCGAGUCAAGUAAACUGCAAGCCAAA-GCCGGGUG------CACUUUUUCACUUUUUUCCAACAGUCUCGCUUU ---...((((.((((....(((((((((....)))))))))...(.((........-)).)))))------))))............................. ( -22.20, z-score = -1.06, R) >droYak2.chr2L 2790687 75 + 22324452 ---CGUAGUGUCGCCAUCAACUUGACU------------------UGCAAGCCAAA-GCCUGGUG------CACUUUUUCACUUUUUU-CAACAGUCUCUCUUU ---...((((.(((((....((((...------------------..)))).....-...)))))------)))).............-............... ( -10.50, z-score = 0.52, R) >droEre2.scaffold_4929 7514421 93 - 26641161 ---CGUAGUGUCGCCAUCAACUUGACUUGCUUGAGUCAAGUAAACUGCAAGCCAAA-GCU-GGUG------CACUUUUUCACAUUUUUUCAAGAGUCCCACUUU ---...((((.((((....(((((((((....)))))))))........(((....-)))-))))------))))................((((.....)))) ( -22.50, z-score = -1.27, R) >droWil1.scaffold_180772 2641236 92 + 8906247 ---CGAGGCA-GGAUAUCAACUUGACUU--UCAAGUCAAGUAAACUGCAAGUAAAAUGCUUUGUG------CGCUUUUUGCUUUGGCCCUACCAUUGACAUUUU ---(((((.(-((......((((((((.--...)))))))).....((.(((((((.((......------.)).)))))))...))))).)).)))....... ( -23.70, z-score = -1.54, R) >droPer1.super_116 87803 91 - 110639 CGCAGGGGCUCUGCCAUCGACUUGACUUCCUCGAGUCAAGUAAACUGCAAGUAAAAUGCUUUGUG------CACUUUUGCUUCCUCGUCCCUGCAUC------- .((((((((...((.....(((((((((....))))))))).....))(((((((((((.....)------)).))))))))....))))))))...------- ( -38.20, z-score = -6.28, R) >dp4.chr4_group4 4627594 91 - 6586962 CGCAGGGGCUCUGCCAUCGACUUGACUUCCUCGAGUCAAGUAAACUGCAAGUAAAAUGCUUUGUG------CACUUUUGCUUCCUCGUCCCUGCAUC------- .((((((((...((.....(((((((((....))))))))).....))(((((((((((.....)------)).))))))))....))))))))...------- ( -38.20, z-score = -6.28, R) >droMoj3.scaffold_6500 23228466 97 + 32352404 ---UUUCGUUUGGUUAU--ACUUGACUU--UGGAGUCGAGCAAACUGCAAGUGAAAUGUUUUGUGUUGCUUCAUUCUUUCUUUUUUGUCCGCUCGCAAAGCUUC ---((((((((((((..--.(((((((.--...)))))))..)))..))))))))).((((((((..((..((............))...)).))))))))... ( -20.90, z-score = -0.56, R) >consensus ___CGUAGUGUCGCCAUCAACUUGACUUGCUCGAGUCAAGUAAACUGCAAGCCAAA_GCUUGGUG______CACUUUUUCACUUUUUUCCAACAGUCUCGCUUU ............((.....(((((((((....))))))))).....))..............(((......))).............................. ( -9.24 = -9.92 + 0.69)
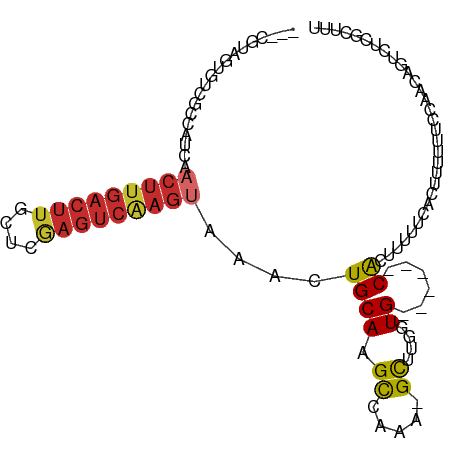
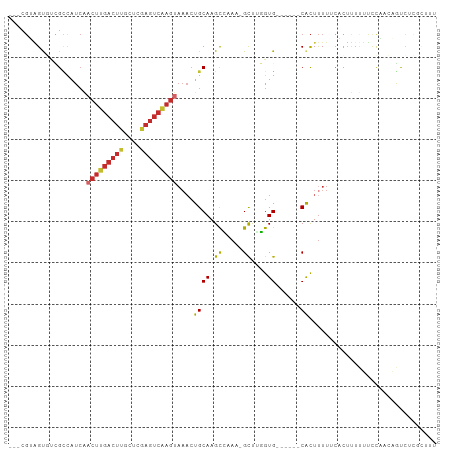
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:40 2011