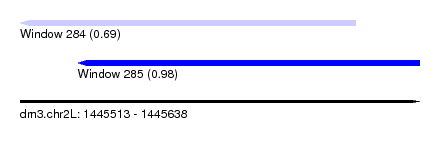
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,445,513 – 1,445,638 |
| Length | 125 |
| Max. P | 0.977585 |

| Location | 1,445,513 – 1,445,618 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 57.97 |
| Shannon entropy | 0.77862 |
| G+C content | 0.56420 |
| Mean single sequence MFE | -26.19 |
| Consensus MFE | -5.70 |
| Energy contribution | -6.42 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.691541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
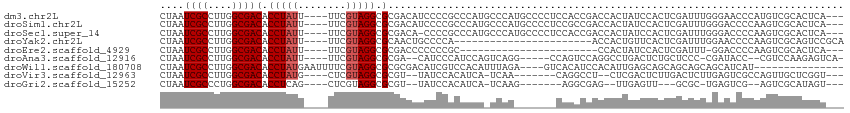
>dm3.chr2L 1445513 105 - 23011544 ----UAUUUUCGUAGGCGCGACAUCCCCGCCCAUGCCCAUGCCCCUCCACCGACCACUAUCCACUCGAUU-UGGGAACCCAUGUCGCA-------CUCACAUCCGUGCGUAUGAGCA ----..........((.(((.......))))).(((.(((((........................(((.-(((....))).)))(((-------(........))))))))).))) ( -26.60, z-score = -1.53, R) >droSim1.chr2L 1420862 105 - 22036055 ----UAUUUUCGUAGGCGCGACAUCCCCGCCCAUGCCCAUGCCCCUCCGCCGACCACUAUCCACUCGAUU-UGGGACCCCAAGUCGCA-------CUCACAUCCGUGCGUAUGAGCA ----..........((.(((.......))))).(((.(((((........................((((-(((....)))))))(((-------(........))))))))).))) ( -29.80, z-score = -2.40, R) >droSec1.super_14 1392993 104 - 2068291 ----UAUUUUCGUAGGCGCGACA-CCCCGCCCAUGCCCAUGCCCCUCCACCGACCACUAUCCACUCGAUU-UGGGACCCCAAGUCGCA-------CUCACAUCCGUGCGUAUGAGCA ----..........((.(((...-...))))).(((.(((((........................((((-(((....)))))))(((-------(........))))))))).))) ( -30.30, z-score = -3.16, R) >droYak2.chr2L 1420762 85 - 22324452 ----UAUUUUCGUAGGCGCAACUGCCCCAAC-----------------------CACUGUUCACUCGAUU-UGGAACCCCAAGUCGCA----GUCCGCACAUCCGUGCGUAUGAGCA ----.......((.((.((....)).)).))-----------------------....((((((((((((-(((....)))))))).)----)).(((((....)))))...)))). ( -27.60, z-score = -3.34, R) >droEre2.scaffold_4929 1490900 81 - 26641161 ----UAUUUUCGUAGGCGCGACCCCCCCGCC-----------------------CACUAUCCACUCGAUU-U-GGACCCCAAGUCGCA-------CUCACAUCCGUGCGUGUGAGCA ----.......((.((((.(......)))))-----------------------.))......(((((((-(-((...)))))))(((-------(.(((....))).))))))).. ( -24.30, z-score = -2.89, R) >droAna3.scaffold_12916 4907313 97 - 16180835 ----UAUUUUCGUAGGCGCGACAUCCCAUCCAGU---------------CAGGCCAGUCCAGGCCUGACU-CUGCUCCCCGAUACCCGUCCAAGAGUCACAUCCGUGCGUAUGAGCA ----....((((((.(((((...........(((---------------((((((......)))))))))-..((((...(((....)))...))))......))))).)))))).. ( -35.00, z-score = -4.30, R) >droWil1.scaffold_180708 9416690 95 + 12563649 UAUGAAUUUUCGUAGGCGCGCGACAUCGUCC------------------ACAUUUAGAGUCACAUCCACAUUGAGCAGCAGCAGCAGC----AUCAUCAUCGUCAUCAUGAUGAGGU (((((....))))).((..(((((.((....------------------.......))))).............((....)).)).))----(((.(((((((....)))))))))) ( -17.10, z-score = 1.10, R) >droVir3.scaffold_12963 7436237 87 - 20206255 ----UAUGCUCGUAGGCGCGUUAUCCACAUC--------------------AUCAACAGGCCUCUCGACUCUUGACUCUUGAGUCGCC----AGUUGCUCGGUUGCCCAGUUGCG-- ----...((.....(((..((.....))...--------------------.......((((...(((((..(((((....)))))..----)))))...))))))).....)).-- ( -18.80, z-score = 1.10, R) >consensus ____UAUUUUCGUAGGCGCGACAUCCCCGCC__________________CCGACCACUAUCCACUCGAUU_UGGGACCCCAAGUCGCA_______CUCACAUCCGUGCGUAUGAGCA ........((((((.(((((...................................................................................))))).)))))).. ( -5.70 = -6.42 + 0.72)
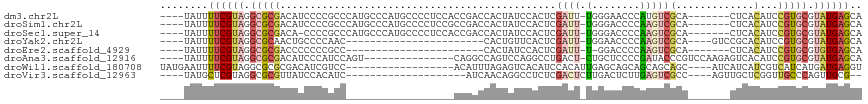
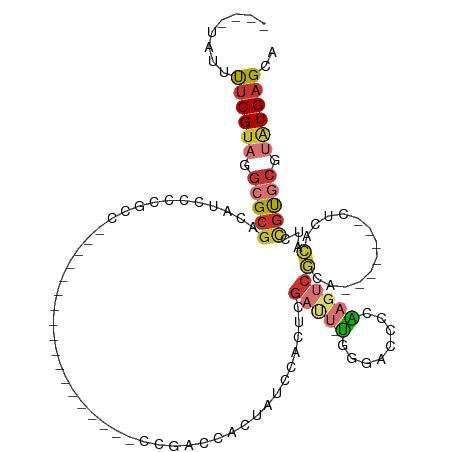
| Location | 1,445,531 – 1,445,638 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 58.49 |
| Shannon entropy | 0.84722 |
| G+C content | 0.57138 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -9.41 |
| Energy contribution | -9.63 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
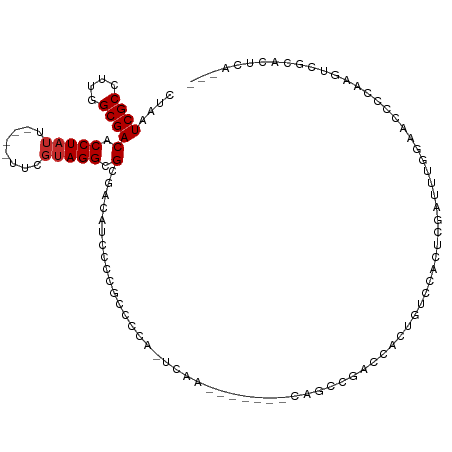
>dm3.chr2L 1445531 107 - 23011544 CUAAUCGCCUUGGCGACACCUAUU----UUCGUAGGCGCGACAUCCCCGCCCAUGCCCAUGCCCCUCCACCGACCACUAUCCACUCGAUUUGGGAACCCAUGUCGCACUCA--- ....((((....))))..(((((.----...))))).(((((((.....((((.((....))........(((...........)))...)))).....))))))).....--- ( -26.40, z-score = -2.06, R) >droSim1.chr2L 1420880 107 - 22036055 CUAAUCGCCUUGGCGACACCUAUU----UUCGUAGGCGCGACAUCCCCGCCCAUGCCCAUGCCCCUCCGCCGACCACUAUCCACUCGAUUUGGGACCCCAAGUCGCACUCA--- ......((.((((((...(((((.----...))))).(((.......))).................)))))).............(((((((....))))))))).....--- ( -26.60, z-score = -1.62, R) >droSec1.super_14 1393011 106 - 2068291 CUAAUCGCCUUGGCGACACCUAUU----UUCGUAGGCGCGACA-CCCCGCCCAUGCCCAUGCCCCUCCACCGACCACUAUCCACUCGAUUUGGGACCCCAAGUCGCACUCA--- ....((((....))))........----...(((((.(((...-...))))).))).............................((((((((....))))))))......--- ( -24.30, z-score = -1.46, R) >droYak2.chr2L 1420780 87 - 22324452 CUAAUCGCCUUGGCGACACCUAUU----UUCGUAGGCGCAACUGCCCCA-----------------------ACCACUGUUCACUCGAUUUGGAACCCCAAGUCGCAGUCCGCA ....((((....))))..(((((.----...))))).((.(((((...(-----------------------((....))).....(((((((....))))))))))))..)). ( -25.30, z-score = -2.65, R) >droEre2.scaffold_4929 1490918 83 - 26641161 CUAAUCGCCUUGGCGACACCUAUU----UUCGUAGGCGCGACCCCCCCGC-----------------------CCACUAUCCACUCGAUUU-GGACCCCAAGUCGCACUCA--- ....((((....))))........----...((.((((.(......))))-----------------------).))........((((((-((...))))))))......--- ( -21.00, z-score = -2.41, R) >droAna3.scaffold_12916 4907331 99 - 16180835 CUAAUCGCCUUGGCGACACCUAUU----UUCGUAGGCGCGA--CAUCCCAUCCAGUCAGG-----CCAGUCCAGGCCUGACUCUGCUCCC-CGAUACC--CGUCCAAGAGUCA- ....((((....))))(.(((((.----...))))).).((--(.........(((((((-----((......)))))))))........-.(((...--.))).....))).- ( -30.10, z-score = -2.33, R) >droWil1.scaffold_180708 9416710 95 + 12563649 CUAAUCGCCUUGGCGACACCUAUGAAUUUUCGUAGGCGCGCGACAUCGUCCACAUUUAGA----GUCACAUCCACAUUGAGCAGCAGCAGCAGCAUCAU--------------- ....((((....))))..(((((((....))))))).(((((((.((...........))----))).............((....)).)).)).....--------------- ( -22.10, z-score = -0.72, R) >droVir3.scaffold_12963 7436249 95 - 20206255 CUAAUCGCCUUGGCGACACCUAUG----CUCGUAGGCGCGU--UAUCCACAUCA-UCAA-------CAGGCCU--CUCGACUCUUGACUCUUGAGUCGCCAGUUGCUCGGU--- ......(((.(((((.(.(((((.----...))))).))))--)).........-.(((-------(.(((..--(((((..........)))))..))).))))...)))--- ( -25.20, z-score = -0.51, R) >droGri2.scaffold_15252 10558226 89 - 17193109 CUAAUCGCCCUGGCGACACCUCAG----CUCGUAGGCGCGU--UAUCCACAUCA-UCAAG-------AGGCGAG--UUGAGUU---GCGC-UGAGUCG--AGUCGCAUAGU--- ....(((.(..((((.((.(((((----(((((.((.....--...))...((.-....)-------).)))))--))))).)---))))-)..).))--)..........--- ( -27.20, z-score = -0.22, R) >consensus CUAAUCGCCUUGGCGACACCUAUU____UUCGUAGGCGCGACAUCCCCGCCCCA_UCAA_______CAGCCGACCACUGUCCACUCGAUUUGGAACCCCAAGUCGCACUCA___ ....((((....))))(.(((((........))))).)............................................................................ ( -9.41 = -9.63 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:36 2011