| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,740,770 – 15,740,863 |
| Length | 93 |
| Max. P | 0.903281 |

| Location | 15,740,770 – 15,740,863 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 64.71 |
| Shannon entropy | 0.71795 |
| G+C content | 0.40687 |
| Mean single sequence MFE | -21.34 |
| Consensus MFE | -9.13 |
| Energy contribution | -8.91 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15740770 93 - 23011544 ---------------UGCUUUUGACGAG-CAGCUUAAAAACAAGAUUCAGGAUAAAGAACCUUGACGGCUUCUCAAGGCCGUACAAUUAUGGGUUUAAAUGCCAAGAGG ---------------..((((((....(-((..(((((..((.((((((((.........))))((((((......))))))..)))).))..))))).))))))))). ( -22.70, z-score = -1.28, R) >droSim1.chr2L 15449773 93 - 22036055 ---------------UGCUUUUGACGAG-CAGUUUAAAAACAAGAUUCAGGAUAAAGAACCUUGACGGCUUCUCAAGGCCGUACAAUUAUGGGUUUAAAUGCCAAGAGG ---------------..((((((....(-((.((((((..((.((((((((.........))))((((((......))))))..)))).))..))))))))))))))). ( -23.00, z-score = -1.55, R) >droSec1.super_3 1949788 93 - 7220098 ---------------UGCUUUUGACGAG-CAGUUUAAAAACAAGAUUCAGGAUAAAGAACUUUGACGGCUUCUCAAGGCCGUACAAUUAUGGGUUUAAAUGCCAAGAGG ---------------..((((((....(-((.((((((..((((.(((........)))))(((((((((......)))))).)))...))..))))))))))))))). ( -21.90, z-score = -1.38, R) >droYak2.chr2L 2703897 93 + 22324452 ---------------UGCUUUUGACGAG-CAGCCUUAAAACCAAAUUCAGAAUAAACAGCCUUGACGGCCUAUUAAGGCCGUACAAUUAUGGGUUUAAAUGCCAAGAGG ---------------(((((.....)))-)).((((....................((...((((((((((....))))))).)))...))(((......)))..)))) ( -24.20, z-score = -1.94, R) >droEre2.scaffold_4929 7440513 91 - 26641161 ---------------UGCUUUUGACGAG-CAGCCUUAAAA--AGAUUCAGGAUCAAGUACCUUGACGGCUUCUCAAGGCCGUACAAUUAUGGGUUUAAAUGCCAAGAGG ---------------..((((((..(((-.((((......--.((((...))))..((......)))))).)))(((.(((((....))))).)))......)))))). ( -21.20, z-score = -0.31, R) >droAna3.scaffold_12916 15560046 93 + 16180835 ---------------UGCUUUCGACGAG-CAGCUCCUUCAACAUAUCCAGGAUUACAAAUACCAACGGCACCUUCAGGCCGUACAAUUAUGGGUUUAUAUGCCAAGAGG ---------------(((((.....)))-))....((((..((((....((..........)).(((((........))))).....))))(((......)))..)))) ( -18.70, z-score = -0.38, R) >dp4.chr4_group4 4517907 94 - 6586962 ---------------UGCCUCCGACGAGCCAGCCUUCACAUAAUAUUCAGGAUUCCUUACCUCAACGGCUUCUCGAGGCCGUACAAUUAUGGGUUUAUAUGACAAGAGG ---------------((.(((....))).))..((((.((((((....(((........)))..(((((((....)))))))...)))))).(((.....)))..)))) ( -20.00, z-score = -0.03, R) >droPer1.super_93 138921 94 - 177177 ---------------UGCCUCCGACGAGCCAGCCUUCACAUAAUAUUCAGGAUUCCUUACCUCAACGGCUUCUCGAGGCCGUACAAUUAUGGGUUUAUAUGACAAGAGG ---------------((.(((....))).))..((((.((((((....(((........)))..(((((((....)))))))...)))))).(((.....)))..)))) ( -20.00, z-score = -0.03, R) >droWil1.scaffold_180772 6123688 105 - 8906247 -GCUGUAAGAAGCAGCAGCAUUUGAAUAUAUUUUCAAUU-AAUAAU-CACU-UUCCAAUUUCAAACGGCUUGUCGAGGCCGUAUAAUUAUGGAUUUAUAUGGCAAGAGG -(((((.....))))).((..(((((......)))))..-......-....-.((((.......(((((((....))))))).......))))........))...... ( -24.34, z-score = -1.85, R) >droVir3.scaffold_12723 4459925 104 - 5802038 UGCUUUAUCGAGCAGC----UCUACAAAUAAAAAAAAUU-AUUGAUAUUCAAAUUUUAACUUCAACGGCAUUUCACGGCCGUACAAUUAUGGAUUUAUAUGCCAAGAGG (((((....))))).(----(((................-.(((.....)))..............(((((.......(((((....)))))......))))).)))). ( -19.52, z-score = -1.89, R) >droMoj3.scaffold_6500 24754162 105 + 32352404 UGCUUUUACGAGCAGC----UCUACUUGAGUUAAAAAUUAAUUGAUAUUCAAUUUAUUACUUCAACGGCGUUUCAAAGCCGUACAAUUAUGGAUCUAUAUGCCAAGAGG (((((....))))).(----(((..(((((.(((.....(((((.....)))))..))).))))).(((((.......(((((....)))))......))))).)))). ( -22.72, z-score = -1.96, R) >droGri2.scaffold_15126 3413089 104 - 8399593 UGCUUUAUCGAGCAGC----UCCACACACUUUAAAAAUU-AUUCAUAUACUAUUCAAAACUUCAACGGCAUUUCAAGGCCGUACAAUUAUGGAUUUUAAUGCCAAGAGG (((((....)))))..----...................-...................((((...((((((......(((((....))))).....))))))..)))) ( -17.80, z-score = -1.84, R) >consensus _______________UGCUUUCGACGAG_CAGCUUAAAAAAAAGAUUCAGGAUUAAGAACCUCAACGGCUUCUCAAGGCCGUACAAUUAUGGGUUUAAAUGCCAAGAGG ...........................................................((((.(((((........))))).......((...........)).)))) ( -9.13 = -8.91 + -0.22)

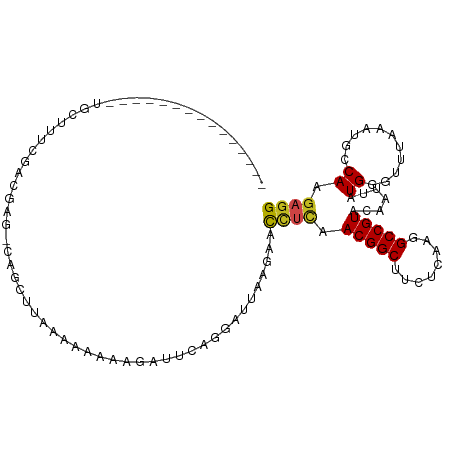
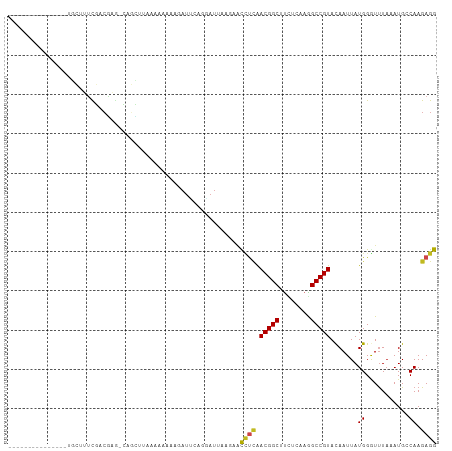
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:33 2011