
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,682,200 – 15,682,324 |
| Length | 124 |
| Max. P | 0.987058 |

| Location | 15,682,200 – 15,682,299 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.41 |
| Shannon entropy | 0.49535 |
| G+C content | 0.50471 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.54 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
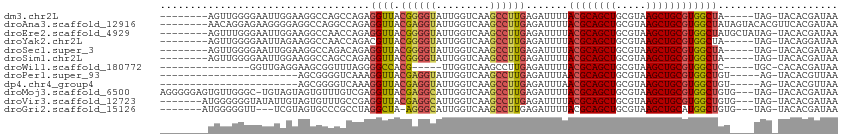
>dm3.chr2L 15682200 99 + 23011544 --------AGUUGGGGAAUUGGAAGGCCAGCCAGAGGUUACGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUA-----UAG-UACACGAUAA --------.((((..(.((((...((((.....((((((.((((((.........)))))).))))))((((((((.....)))))))))))).-----)))-).).)))).. ( -31.30, z-score = -1.07, R) >droAna3.scaffold_12916 15546640 105 - 16180835 --------AACAGGAGAAGGGGAGGCCAGGCCAGAGGUUACGAGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUAUAGUACACGUUCACGAUAA --------.......(((.(....((..((((.((((((.((((((.........)))))).))))))((((((((.....))))))))))))...))...).)))....... ( -33.50, z-score = -1.53, R) >droEre2.scaffold_4929 7426111 104 + 26641161 --------AGUUUGGGAAUUGGAAGGCCAACCAGAGGUUACGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUAUGCUAUAG-UACACGAUAA --------..(((((...((((....))))))))).....((..((((((..((((((..........((((((((.....)))))))))))).))...)))-))).)).... ( -30.40, z-score = -0.50, R) >droYak2.chr2L 2688553 99 - 22324452 --------AGUUGGGGAAUUAGAAGGCCAACCAGACGUUACGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUA-----UAG-UACAGGAUAA --------((((..((((((..(((((..(((((((........)).)))))...)))))..))))))((((((((.....)))))))))))).-----...-.......... ( -29.20, z-score = -0.93, R) >droSec1.super_3 1935100 99 + 7220098 --------AGUUGGGGAAUUGGAAGGCCAGACAGAGGUUACGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUA-----UAG-UACACGAUAA --------.((((..(.((((...((((.....((((((.((((((.........)))))).))))))((((((((.....)))))))))))).-----)))-).).)))).. ( -31.30, z-score = -1.87, R) >droSim1.chr2L 15435174 99 + 22036055 --------AGUUGGGGAAUUGGAAGGCCAGCCAGAGGUUACGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUA-----UAG-UACACGAUAA --------.((((..(.((((...((((.....((((((.((((((.........)))))).))))))((((((((.....)))))))))))).-----)))-).).)))).. ( -31.30, z-score = -1.07, R) >droWil1.scaffold_180772 6108147 87 + 8906247 ---------------GGUUGAGGAAGCGGUUUAGGGGCCACG-----UUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUC-----UGC-CACACGAUAA ---------------(((.(((.....(((((...((((...-----..)))))))))........((((((((((.....)))))))))))))-----.))-)......... ( -31.60, z-score = -1.03, R) >droPer1.super_93 114379 84 + 177177 -----------------------AGCGGGGUCAAAGGUUACGAGGUAUUGGUCAAGCCUUGAGAUUUAACGCAGCUGCGUAAGCUGCGUGGCUGU-----AG-UACACGUUAA -----------------------((((.((((..(((((.((((((.........)))))).))))).((((((((.....))))))))))))(.-----..-..).)))).. ( -26.60, z-score = -0.92, R) >dp4.chr4_group4 4493593 84 + 6586962 -----------------------AGCGGGGUCAAAGGUUACGAGGUAUUGGUCAAGCCUUGAGAUUUAACGCAGCUGCGUAAGCUGCGUGGCUGU-----AG-UACACGUUAA -----------------------((((.((((..(((((.((((((.........)))))).))))).((((((((.....))))))))))))(.-----..-..).)))).. ( -26.60, z-score = -0.92, R) >droMoj3.scaffold_6500 24731854 108 - 32352404 AGGGGGAGUGUUGGGC-UGUAGUAGUGUUUGUCGAGGUUACGAGGCAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUGUG---UAG-UACACGAUAA .......((((.((((-(..(.(((((((..(((......)))))))))).)..))))).......(((((((((..(((.....)))..))))))---)))-.))))..... ( -37.20, z-score = -1.77, R) >droVir3.scaffold_12723 4441721 102 + 5802038 -------AUGGGGGGUAUAUUGUAGUGUUUGCCGAGGUUACGAGGCAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUGUG---UAG-UACACGAUAA -------..........((((((.(((......((((((.((((((.........)))))).))))))(((((((..(((.....)))..))))))---)..-))))))))). ( -34.50, z-score = -1.52, R) >droGri2.scaffold_15126 3394661 98 + 8399593 -------AUGGGGGUU---UCGUAGUGCCCGCCUAGGCUA-AGGGCAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCAUGGCUGUG---UAG-UACACGAUAA -------.(.((((((---(..((((((((((....))..-.))))))))...))))))).)....((((((((((((....)).....)))))))---)))-.......... ( -35.30, z-score = -0.85, R) >consensus ________AGUUGGGGAAUUGGAAGGCCAGCCAGAGGUUACGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUAU____UAG_UACACGAUAA ...................................((((.((((((.........)))))).......((((((((.....)))))))))))).................... (-20.62 = -20.54 + -0.08)

| Location | 15,682,200 – 15,682,299 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.41 |
| Shannon entropy | 0.49535 |
| G+C content | 0.50471 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.31 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15682200 99 - 23011544 UUAUCGUGUA-CUA-----UAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCGUAACCUCUGGCUGGCCUUCCAAUUCCCCAACU-------- ..........-...-----.....((((((((.....))))))))........((((((.(.(((...............)))).))))))..............-------- ( -21.86, z-score = -1.22, R) >droAna3.scaffold_12916 15546640 105 + 16180835 UUAUCGUGAACGUGUACUAUAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCUCGUAACCUCUGGCCUGGCCUCCCCUUCUCCUGUU-------- ....((....)).........(((((((((((.....)))))))).........(((((.((....(((...)))...)).))))))))................-------- ( -24.80, z-score = -1.17, R) >droEre2.scaffold_4929 7426111 104 - 26641161 UUAUCGUGUA-CUAUAGCAUAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCGUAACCUCUGGUUGGCCUUCCAAUUCCCAAACU-------- .....((((.-.....))))....((((((((.....))))))))........((((((.(((((...............)))))))))))..............-------- ( -24.36, z-score = -1.63, R) >droYak2.chr2L 2688553 99 + 22324452 UUAUCCUGUA-CUA-----UAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCGUAACGUCUGGUUGGCCUUCUAAUUCCCCAACU-------- ..........-...-----.....((((((((.....))))))))........((((((.(((((..((........)).)))))))))))..............-------- ( -23.00, z-score = -1.75, R) >droSec1.super_3 1935100 99 - 7220098 UUAUCGUGUA-CUA-----UAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCGUAACCUCUGUCUGGCCUUCCAAUUCCCCAACU-------- ..........-...-----.....((((((((.....))))))))........((((((.(((..................))).))))))..............-------- ( -21.57, z-score = -2.19, R) >droSim1.chr2L 15435174 99 - 22036055 UUAUCGUGUA-CUA-----UAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCGUAACCUCUGGCUGGCCUUCCAAUUCCCCAACU-------- ..........-...-----.....((((((((.....))))))))........((((((.(.(((...............)))).))))))..............-------- ( -21.86, z-score = -1.22, R) >droWil1.scaffold_180772 6108147 87 - 8906247 UUAUCGUGUG-GCA-----GAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAA-----CGUGGCCCCUAAACCGCUUCCUCAACC--------------- .....((.((-(..-----(((((((((((((.....))))))))..........)))))..))))-----)((((........))))..........--------------- ( -27.00, z-score = -2.05, R) >droPer1.super_93 114379 84 - 177177 UUAACGUGUA-CU-----ACAGCCACGCAGCUUACGCAGCUGCGUUAAAUCUCAAGGCUUGACCAAUACCUCGUAACCUUUGACCCCGCU----------------------- ...(((.(((-..-----..((((((((((((.....))))))))..........)))).......)))..)))................----------------------- ( -17.50, z-score = -0.87, R) >dp4.chr4_group4 4493593 84 - 6586962 UUAACGUGUA-CU-----ACAGCCACGCAGCUUACGCAGCUGCGUUAAAUCUCAAGGCUUGACCAAUACCUCGUAACCUUUGACCCCGCU----------------------- ...(((.(((-..-----..((((((((((((.....))))))))..........)))).......)))..)))................----------------------- ( -17.50, z-score = -0.87, R) >droMoj3.scaffold_6500 24731854 108 + 32352404 UUAUCGUGUA-CUA---CACAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUGCCUCGUAACCUCGACAAACACUACUACA-GCCCAACACUCCCCCU .....((((.-...---.......((((((((.....))))))))..........((((...........(((......))).............)-)))..))))....... ( -21.45, z-score = -2.01, R) >droVir3.scaffold_12723 4441721 102 - 5802038 UUAUCGUGUA-CUA---CACAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUGCCUCGUAACCUCGGCAAACACUACAAUAUACCCCCCAU------- .....(((..-...---))).(((((((((((.....)))))))).........((((.........)))).........))).......................------- ( -25.60, z-score = -3.20, R) >droGri2.scaffold_15126 3394661 98 - 8399593 UUAUCGUGUA-CUA---CACAGCCAUGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUGCCCU-UAGCCUAGGCGGGCACUACGA---AACCCCCAU------- ...(((((..-...---((.((((((((((((.....))))))))..........)))))).....(((((.-..((....))))))).)))))---.........------- ( -28.70, z-score = -1.68, R) >consensus UUAUCGUGUA_CUA____AUAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCGUAACCUCUGGCUGGCCUUCCAAUUCCCCAACU________ ....................((((((((((((.....))))))))..........))))...................................................... (-15.38 = -15.31 + -0.08)
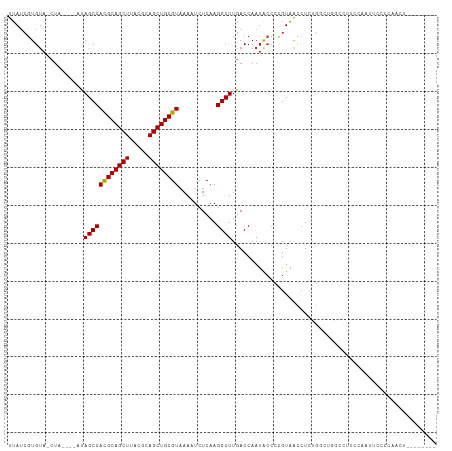
| Location | 15,682,232 – 15,682,324 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Shannon entropy | 0.35839 |
| G+C content | 0.47395 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -23.98 |
| Energy contribution | -24.32 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.987058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
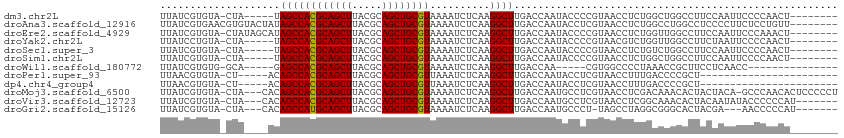
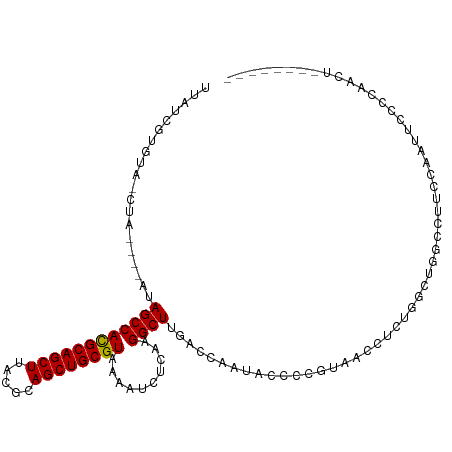
>dm3.chr2L 15682232 92 + 23011544 CGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUAU-----A-GUACACGAUAA-ACCGUUAUUAUGUGCCAUGUGCCCC------------ .(((((((((((..((((..........((((((((.....))))))))))))((-----(-(((.(((....-..)))))))))..)))).)))))))------------ ( -36.30, z-score = -3.25, R) >droAna3.scaffold_12916 15546672 98 - 16180835 CGAGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUAUAGUACACGUUCACGAUAA-ACCGUUAUUAUGUGCUAUAUACACG------------ ((((((.........)))))).........((((((.....))))))(((..(((((((((.....(((....-..))).....)))))))))..))).------------ ( -32.60, z-score = -2.46, R) >droEre2.scaffold_4929 7426143 97 + 26641161 CGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUAUGCUAUA-GUACACGAUAA-ACCGUUAUUAUGUGCCAUGUGCCCC------------ .(((((((((((..((((..........((((((((.....))))))))))))....((((-(((.(((....-..)))))))))).)))).)))))))------------ ( -37.10, z-score = -2.85, R) >droYak2.chr2L 2688585 91 - 22324452 CGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUAU-----A-GUACAGGAUAA-ACCGUUAUUAUGUGCCCUGUGCAC------------- ((((((((......((((..........((((((((.....))))))))))))((-----(-(((..((....-.))..)))))))))))))).....------------- ( -31.40, z-score = -1.60, R) >droSec1.super_3 1935132 92 + 7220098 CGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUAU-----A-GUACACGAUAA-GCCGUUAUUAUGUGCCAUGUGCCCC------------ .(((((((((((..((((..........((((((((.....))))))))))))((-----(-(((.(((....-..)))))))))..)))).)))))))------------ ( -36.30, z-score = -2.87, R) >droSim1.chr2L 15435206 92 + 22036055 CGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUAU-----A-GUACACGAUAA-ACCGUUAUUAUGUGCCAUGUGCCCC------------ .(((((((((((..((((..........((((((((.....))))))))))))((-----(-(((.(((....-..)))))))))..)))).)))))))------------ ( -36.30, z-score = -3.25, R) >droWil1.scaffold_180772 6108172 89 + 8906247 -----CGUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUCUG------CCACACGAUAA-ACCGUUAUUAUGUGCUACCUAUAUGUA---------- -----(((((((..((((..........((((((((.....))))))))))))..)------))).)))....-........((((((.....))))))..---------- ( -26.00, z-score = -1.12, R) >droPer1.super_93 114396 90 + 177177 CGAGGUAUUGGUCAAGCCUUGAGAUUUAACGCAGCUGCGUAAGCUGCGUGGCUGU-----A-GUACACGUUAA-GCCGUUAUUAUGUGCCAUGUAUA-------------- ((((((.........)))))).......((((((((.....))))))))((((((-----(-(((.(((....-..)))))))))).))).......-------------- ( -29.50, z-score = -1.39, R) >dp4.chr4_group4 4493610 90 + 6586962 CGAGGUAUUGGUCAAGCCUUGAGAUUUAACGCAGCUGCGUAAGCUGCGUGGCUGU-----A-GUACACGUUAA-GCCGUUAUUAUGUGCCAUGUAUA-------------- ((((((.........)))))).......((((((((.....))))))))((((((-----(-(((.(((....-..)))))))))).))).......-------------- ( -29.50, z-score = -1.39, R) >droMoj3.scaffold_6500 24731893 99 - 32352404 CGAGGCAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUGUG---UA-GUACACGAUAAUGCCGUUAUUAUAUACUAUACGA-AUACACA------- ((((((.........)))))).....((((((((((.....)))))))))).((((---((-(((.(..(((((...)))))..).))))))))).-.......------- ( -33.30, z-score = -2.42, R) >droVir3.scaffold_12723 4441754 99 + 5802038 CGAGGCAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUGUG---UA-GUACACGAUAAUACCGUUAUUAUGUACUACACGA-AUACCAA------- ((((((.........)))))).....((((((((((.....)))))))))).((((---((-(((((..(((((...)))))..))))))))))).-.......------- ( -41.20, z-score = -5.08, R) >droGri2.scaffold_15126 3394691 106 + 8399593 -AGGGCAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCAUGGCUGUG---UA-GUACACGAUAAUACCGUUAUUAUGUAUCGCACUAUACACCACAUGUAUA -...((((((((..((((.((((...((((((....)))))).)).)).))))(((---..-(((((..(((((...)))))..)))))..))).....)))).))))... ( -34.20, z-score = -1.79, R) >consensus CGGGGUAUUGGUCAAGCCUUGAGAUUUUACGCAGCUGCGUAAGCUGCGUGGCUAU_____A_GUACACGAUAA_ACCGUUAUUAUGUGCCAUGUGCACC____________ ((((((.........)))))).....((((((((((.....))))))))))...........(((((..((((.....))))..)))))...................... (-23.98 = -24.32 + 0.34)

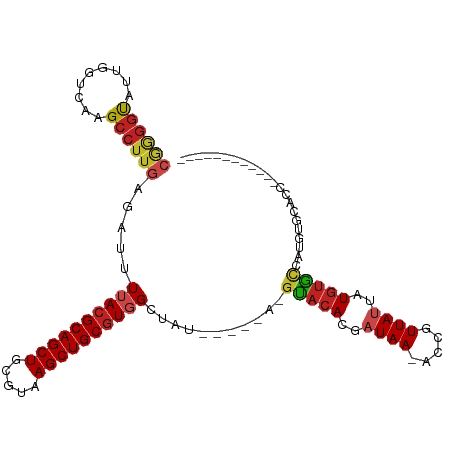

| Location | 15,682,232 – 15,682,324 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Shannon entropy | 0.35839 |
| G+C content | 0.47395 |
| Mean single sequence MFE | -27.91 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.78 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15682232 92 - 23011544 ------------GGGGCACAUGGCACAUAAUAACGGU-UUAUCGUGUAC-U-----AUAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCG ------------((((...((((.(((..((((....-))))..))).)-)-----))((((((((((((.....))))))))..........)))).........)))). ( -29.20, z-score = -2.41, R) >droAna3.scaffold_12916 15546672 98 + 16180835 ------------CGUGUAUAUAGCACAUAAUAACGGU-UUAUCGUGAACGUGUACUAUAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCUCG ------------.((((.(((((.((((....((((.-...))))....)))).)))))(((((((((((.....))))))))..........))).......)))).... ( -25.70, z-score = -1.36, R) >droEre2.scaffold_4929 7426143 97 - 26641161 ------------GGGGCACAUGGCACAUAAUAACGGU-UUAUCGUGUAC-UAUAGCAUAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCG ------------((((...((((.(((..((((....-))))..))).)-)))..((.((((((((((((.....))))))))..........)))))).......)))). ( -29.90, z-score = -2.09, R) >droYak2.chr2L 2688585 91 + 22324452 -------------GUGCACAGGGCACAUAAUAACGGU-UUAUCCUGUAC-U-----AUAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCG -------------((((.....))))........(((-(...(((....-.-----......((((((((.....)))))))).........)))...))))......... ( -25.05, z-score = -1.77, R) >droSec1.super_3 1935132 92 - 7220098 ------------GGGGCACAUGGCACAUAAUAACGGC-UUAUCGUGUAC-U-----AUAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCG ------------((((...((((.(((..((((....-))))..))).)-)-----))((((((((((((.....))))))))..........)))).........)))). ( -29.20, z-score = -2.28, R) >droSim1.chr2L 15435206 92 - 22036055 ------------GGGGCACAUGGCACAUAAUAACGGU-UUAUCGUGUAC-U-----AUAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCG ------------((((...((((.(((..((((....-))))..))).)-)-----))((((((((((((.....))))))))..........)))).........)))). ( -29.20, z-score = -2.41, R) >droWil1.scaffold_180772 6108172 89 - 8906247 ----------UACAUAUAGGUAGCACAUAAUAACGGU-UUAUCGUGUGG------CAGAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAACG----- ----------........(((..((((..((((....-))))..)))).------..(((((((((((((.....))))))))..........))))).)))....----- ( -26.70, z-score = -2.11, R) >droPer1.super_93 114396 90 - 177177 --------------UAUACAUGGCACAUAAUAACGGC-UUAACGUGUAC-U-----ACAGCCACGCAGCUUACGCAGCUGCGUUAAAUCUCAAGGCUUGACCAAUACCUCG --------------......((((........(((..-....)))....-.-----...)))).((((((.....))))))(((((..(....)..))))).......... ( -21.09, z-score = -1.19, R) >dp4.chr4_group4 4493610 90 - 6586962 --------------UAUACAUGGCACAUAAUAACGGC-UUAACGUGUAC-U-----ACAGCCACGCAGCUUACGCAGCUGCGUUAAAUCUCAAGGCUUGACCAAUACCUCG --------------......((((........(((..-....)))....-.-----...)))).((((((.....))))))(((((..(....)..))))).......... ( -21.09, z-score = -1.19, R) >droMoj3.scaffold_6500 24731893 99 + 32352404 -------UGUGUAU-UCGUAUAGUAUAUAAUAACGGCAUUAUCGUGUAC-UA---CACAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUGCCUCG -------(((((((-(.....)))))))).....((((((.(((((...-..---)))((((((((((((.....))))))))..........)))).))..))))))... ( -29.50, z-score = -2.44, R) >droVir3.scaffold_12723 4441754 99 - 5802038 -------UUGGUAU-UCGUGUAGUACAUAAUAACGGUAUUAUCGUGUAC-UA---CACAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUGCCUCG -------..(((((-(.((((((((((..((((.....))))..)))))-))---)))((((((((((((.....))))))))..........)))).....))))))... ( -38.40, z-score = -5.13, R) >droGri2.scaffold_15126 3394691 106 - 8399593 UAUACAUGUGGUGUAUAGUGCGAUACAUAAUAACGGUAUUAUCGUGUAC-UA---CACAGCCAUGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUGCCCU- ..(((..(((((((.(((((((....((((((....))))))..)))))-))---.))).))))((((((.....))))))))).........(((.........)))..- ( -29.90, z-score = -1.03, R) >consensus ____________GGUGCACAUGGCACAUAAUAACGGU_UUAUCGUGUAC_U_____AUAGCCACGCAGCUUACGCAGCUGCGUAAAAUCUCAAGGCUUGACCAAUACCCCG ........................((((.((((.....)))).))))...........((((((((((((.....))))))))..........)))).............. (-16.60 = -16.78 + 0.18)

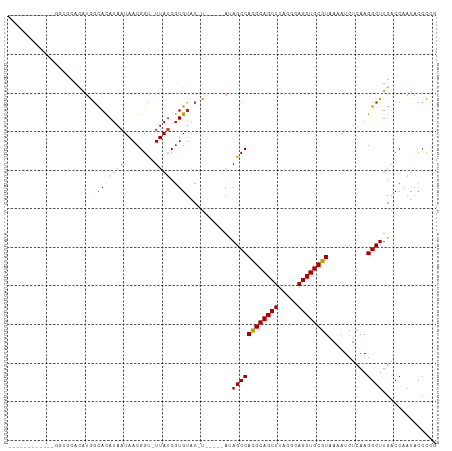
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:28 2011