
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,652,634 – 15,652,736 |
| Length | 102 |
| Max. P | 0.572950 |

| Location | 15,652,634 – 15,652,736 |
|---|---|
| Length | 102 |
| Sequences | 15 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.47 |
| Shannon entropy | 0.58717 |
| G+C content | 0.57087 |
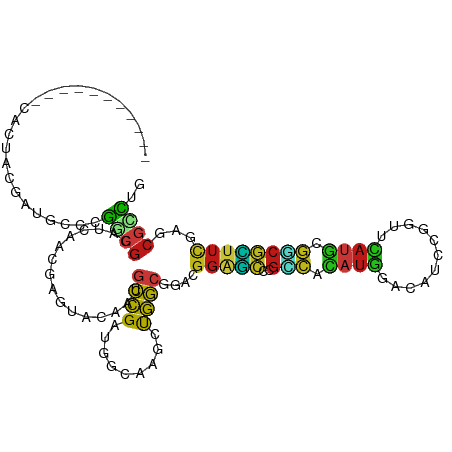
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -15.71 |
| Energy contribution | -14.90 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 2.05 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.572950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15652634 102 - 23011544 -----------CACUACGAUGCCCGGCGGAUCAACGAAUACAAUGCCGAUGGCAAGCUGGCGGACGGAGCCCGCCACAUGGACAUCCGGUUCAUGCGGCGCUUCGAGCGCCUG -----------........(((((((((...............)))))..))))....((((..((((((..(((.(((((((.....))))))).)))))))))..)))).. ( -41.36, z-score = -1.69, R) >droPer1.super_93 75556 105 - 177177 --------CAACACUACGAUGCGAGGCGUAUCAACGAGUAUAAUGCCGAUGGCAAGCUGGCGGACGGAGCCCGUCAUAUGGAUAUACGGUUCAUGCGACGCUUCGAACGCCUC --------..............(((((((.((..(.(((....((((...)))).))).)..))((((((..(((.(((((((.....))))))).))))))))).))))))) ( -35.80, z-score = -1.39, R) >dp4.chr4_group4 4453322 105 - 6586962 --------CAACACUACGAUGCGAGGCGUAUCAACGAGUAUAAUGCCGAUGGCAAGCUGGCGGACGGAGCCCGUCAUAUGGAUAUACGGUUCAUGCGACGCUUCGAACGCCUC --------..............(((((((.((..(.(((....((((...)))).))).)..))((((((..(((.(((((((.....))))))).))))))))).))))))) ( -35.80, z-score = -1.39, R) >droAna3.scaffold_12916 15522917 102 + 16180835 -----------CACUACGAUGCCCGGCGGAUUAACGAGUACAACGCCGACGGCAAGCUGGCGGACGGAGCCCGGCACAUGGACAUCCGGUUCAUGCGGCGCUUUGAGCGCCUG -----------........(((((((((...............)))))..))))....((((..(((((((((...(((((((.....)))))))))).))))))..)))).. ( -39.56, z-score = -1.02, R) >droEre2.scaffold_4929 7398257 102 - 26641161 -----------CACUACGAUGCCCGGCGGAUCAAUGAGUACAAUGCCGAUGGCAAGCUGGCGGACGGAGCCCGCCACAUGGACAUCCGGUUCAUGCGGCGCUUCGAGCGGCUG -----------........(((((((((...............)))))..))))(((((.(...((((((..(((.(((((((.....))))))).))))))))).)))))). ( -39.66, z-score = -0.64, R) >droYak2.chr2L 2656642 102 + 22324452 -----------CACUACGAUGCCCGGCGGAUCAACGAAUACAAUGCCGAUGGGAAGCUGGCGGACGGAGCCCGCCACAUGGACAUCCGGUUCAUGCGGCGCUUCGAGCGCCUG -----------......(...((((.(((................))).))))...).((((..((((((..(((.(((((((.....))))))).)))))))))..)))).. ( -38.89, z-score = -0.96, R) >droSec1.super_3 1907359 102 - 7220098 -----------CACUACGAUGCCCGGCGGAUCAACGAAUACAAUGCCGAUGGCAAGCUGGCGGACGGAGCCCGCCACAUGGACAUCCGGUUCAUGCGGCGCUUCGAGCGUCUG -----------......(((((((((((......)........((((...)))).)))))....((((((..(((.(((((((.....))))))).))))))))).))))).. ( -40.10, z-score = -1.37, R) >droSim1.chr2L 15405886 102 - 22036055 -----------CACUACGAUGCCCGGCGGAUCAACGAAUACAAUGCCGAUGGCAAGCUGGCGGACGGAGCCCGCCACAUGGACAUCCGGUUCAUGCGGCGCUUCGAGCGUCUG -----------......(((((((((((......)........((((...)))).)))))....((((((..(((.(((((((.....))))))).))))))))).))))).. ( -40.10, z-score = -1.37, R) >droWil1.scaffold_180772 6060829 102 - 8906247 -----------CACUACGAUGCUCGACGUAUCAAUGAGUAUAAUGCCGAUGGCAAGUUGGCGGACGGAGCUCGCCACAUGGAUAUACGAUUCAUGAGGCGCUUCGAACGCCUG -----------.(((..(((((.....)))))....)))....((((...))))....((((..((((((..(((.(((((((.....))))))).)))))))))..)))).. ( -37.90, z-score = -2.94, R) >droVir3.scaffold_12723 4391422 102 - 5802038 -----------CAUUACGAUGCAAGACGCAUCAAUGAGUAUAAUGCCGAUGGCAAGUUGGCGGACGGAGCACGCCACAUGGAUAUUCGAUUCAUGCGGCGCUUCGAGCGCCUG -----------......(((((.....)))))...........((((...))))....((((..((((((..(((.(((((((.....))))))).)))))))))..)))).. ( -39.00, z-score = -2.58, R) >droMoj3.scaffold_6500 24693747 102 + 32352404 -----------CAUUACGAUGCAAGGCGCAUCAAUGAGUAUAAUGCCGAUGGCAAAUUGGCGGACGGAGCGCGUCACAUGGACAUACGAUUCAUGCGUCGCUUCGAGCGUCUG -----------............((((((..............(((((((.....)))))))..(((((((((...(((((((....).)))))))).))))))).)))))). ( -33.90, z-score = -0.79, R) >droGri2.scaffold_15126 3357739 102 - 8399593 -----------CAUUACGAUGCGAGACGCAUCAAUGAGUACAAUGCCGAUGGCAAGUUGGCGGACGGAGCUCGCCACAUGGAUAUACGAUUCAUGCGGCGCUUCGAGCGUCUG -----------......(((((..((.((......((((.(..(((((((.....)))))))....).))))(((.(((((((.....))))))).))))).))..))))).. ( -39.70, z-score = -2.55, R) >anoGam1.chr3R 37575839 104 - 53272125 ---------CAUUCUACGACGCGCGCCGUAUCAACGAGUACCAGUCCGAUGGCCGGCUGGCGGAAGGGUCGCGGCAGAUGCUGCUCCGCCAUAUGCGUCGGUUCGAGGGCUAC ---------.......(((((((.((((((((.((........))..))))..))))(((((((......(((((....))))))))))))..)))))))(((....)))... ( -38.80, z-score = 0.63, R) >apiMel3.Group11 14092 102 - 12576330 -----------UAUUACGCUUCUCAACGUUUCGACUCUUCUUCGGGUGAAGGAAAAGCAUCGAUAACCGUACAAGAAACUUUCCUUUCAUCGAAUCGACAUUUCGAUCAUUUG -----------....................(((.......)))((((((((.((((..((.............))..)))).))))))))((.((((....))))))..... ( -15.42, z-score = 0.98, R) >triCas2.chrUn_76 27991 113 + 324113 AACCCUUUCGUUAUUAUAACGCCAAAAGACUGAAUUCGUUUGGUCCUGAUGGACGACCAGUCGAGGGCACGAGAGAAAUGAUGCUAACGCCGAAUCCUCAUUUCGACCAUUUA ....(((((((((...))))(((....(((((.....(((..((((....))))))))))))...)))..)))))(((((..((....))((((.......))))..))))). ( -29.60, z-score = -1.66, R) >consensus ___________CACUACGAUGCCCGGCGGAUCAACGAGUACAAUGCCGAUGGCAAGCUGGCGGACGGAGCCCGCCACAUGGACAUCCGGUUCAUGCGGCGCUUCGAGCGCCUG ........................((((................((((.........))))....(((((..(((.((((...........)))).))))))))...)))).. (-15.71 = -14.90 + -0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:24 2011