| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,650,354 – 15,650,445 |
| Length | 91 |
| Max. P | 0.759276 |

| Location | 15,650,354 – 15,650,445 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 61.32 |
| Shannon entropy | 0.72189 |
| G+C content | 0.50178 |
| Mean single sequence MFE | -25.55 |
| Consensus MFE | -10.49 |
| Energy contribution | -10.65 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.694817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
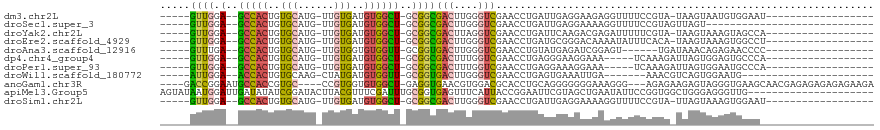
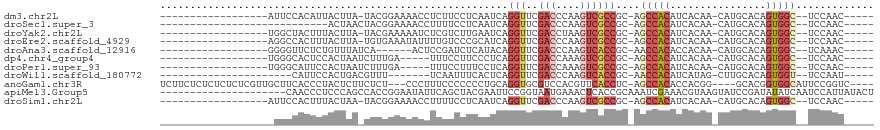
>dm3.chr2L 15650354 91 + 23011544 -----GUUGGA--GCCACUGUGCAUG-UUGUGAUGUGGCU-GCGGCGACUUGGGUCGAACCUGAUUGAGGAAGAGGUUUUCCGUA-UAAGUAAUGUGGAAU------------------ -----((((.(--(((((...((...-..))...))))))-.))))(((....)))((((((...........))))))((((((-(.....)))))))..------------------ ( -26.80, z-score = -1.53, R) >droSec1.super_3 1905058 82 + 7220098 -----GUUGGA--GCCACUGUGCAUG-UUGUGAUGUGGCU-GCGGCGACUUGGGUCGAACCUGAUUGAGGAAAAGGUUUUCCGUAGUUAGU---------------------------- -----((((.(--(((((...((...-..))...))))))-.))))(((....)))....(((((((.(((((....))))).))))))).---------------------------- ( -29.10, z-score = -2.59, R) >droYak2.chr2L 2654299 91 - 22324452 -----GUUGGA--GCCACUGUGCAUG-UUGUGAUGUGGCU-GCGGCGACUUAGGUCGAACCUGAUUCAAGACGAGAUUUUUCGUA-UAAGUAAAGUAGCCA------------------ -----...(.(--(((((...((...-..))...))))))-.)(((((.(((((.....))))).))...(((((....))))).-...........))).------------------ ( -28.10, z-score = -1.68, R) >droEre2.scaffold_4929 7395769 91 + 26641161 -----GUUGGA--GCCACUGUGCAUG-UUGUGAUGUGGCU-GCGGCGACUUGGGUCGAACCUGAUGCGGGACAAAAUAUUUCACA-UAAGUAAAGUGGCCU------------------ -----...((.--((((((.(((.((-(.((((.(((((.-...))..((((.((((....)))).))))......))).)))))-)).))).))))))))------------------ ( -27.20, z-score = -0.51, R) >droAna3.scaffold_12916 15520855 86 - 16180835 -----GUUUGA--GCCACUGUGCAUG-UUGUGGUGUGGUU-GCGGUGACUUGGGUCGAACCUGUAUGAGAUCGGAGU------UGAUAAACAGAGAACCCC------------------ -----((((((--(((((..(((...-..)))..))))))-.....(((((.((((............)))).))))------)..)))))..........------------------ ( -20.00, z-score = 0.57, R) >dp4.chr4_group4 4451227 87 + 6586962 -----GUUGGA--GCCACUGUGCAUG-UUGUGAUGUGGCU-GCGGCGACUUUGGUCGAACCUGAGGGAAGGAAA-----UCAAAGAUUAGUGGAGUGCCCA------------------ -----((((.(--(((((...((...-..))...))))))-.))))..(((((((....(((......)))..)-----)))))).....(((.....)))------------------ ( -26.10, z-score = -0.56, R) >droPer1.super_93 73461 87 + 177177 -----GUUGGA--GCCACUGUGCAUG-UUGUGAUGUGGCU-GCGGCGACUUUGGUCGAACCUGAGGAAAGGAAA-----UCAAAGAUUAGUGGAAUGCCCA------------------ -----((((.(--(((((...((...-..))...))))))-.))))..(((((((....(((......)))..)-----)))))).....(((.....)))------------------ ( -26.10, z-score = -1.08, R) >droWil1.scaffold_180772 6058748 81 + 8906247 -----AUUGGA--ACCACUGUGCAAG-CUAUGAUGUGGUU-GCGGUGACUUGGGUCGAACCUGAGUGAAAUUGA-------AAACGUCAGUGGAAUG---------------------- -----......--.(((((((((((.-(........).))-))).(.(((..((.....))..))).)......-------......))))))....---------------------- ( -23.50, z-score = -2.21, R) >anoGam1.chr3R 37573260 107 + 53272125 ----GACCGGAAUGCCACCGUGC----CCGUGGUGUGGCU-GAGGUGAACGUGGACGCACCUGCAGGGGGGGAAAGGG---AGAGAAGAGUAGGGUGAAGCAACGAGAGAGAGAGAAGA ----.(((((....)).((.(.(----(.((((((((..(-.(.(....).).).)))))).)).)).).))......---............))).......(....).......... ( -23.60, z-score = 1.13, R) >apiMel3.Group5 9631758 98 + 13386189 AGUAUAAUGGAUUGAUAUAUCGGAUACUUACGUUUCGAUUUGCGGUGAGUUUCAUUACCGGAAUUCGUAGCUGAAUAUUCCGGUGGCUGGGAGGGUUG--------------------- .................(((((((((.(((.(((.(((.((.((((((......)))))).)).))).)))))).)).))))))).............--------------------- ( -23.50, z-score = -0.17, R) >droSim1.chr2L 15403635 91 + 22036055 -----GUUGGA--GCCACUGUGCAUG-UUGUGAUGUGGCU-GCGGCGACUUGGGUCGAACCUGAUUGAGGAAAAGGUUUUCCGUA-UUAGUAAAGUGGAAU------------------ -----((((.(--(((((...((...-..))...))))))-.))))(((....)))....(((((...(((((....)))))..)-))))...........------------------ ( -27.10, z-score = -1.72, R) >consensus _____GUUGGA__GCCACUGUGCAUG_UUGUGAUGUGGCU_GCGGCGACUUGGGUCGAACCUGAUUGAGGAAAAAGUUUUCCGUA_UUAGUAGAGUGGCC___________________ ........((...(((((..((((....))))..)))))......((((....))))..)).......................................................... (-10.49 = -10.65 + 0.17)
| Location | 15,650,354 – 15,650,445 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 61.32 |
| Shannon entropy | 0.72189 |
| G+C content | 0.50178 |
| Mean single sequence MFE | -18.03 |
| Consensus MFE | -5.88 |
| Energy contribution | -6.58 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.759276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
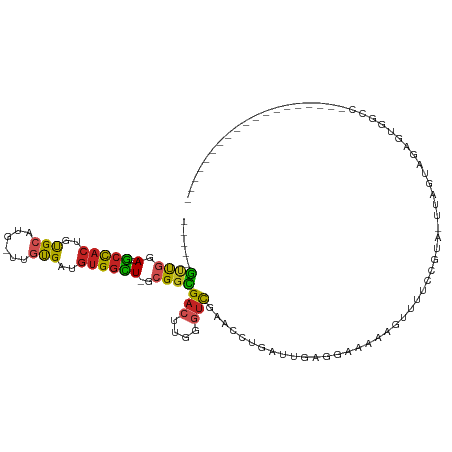
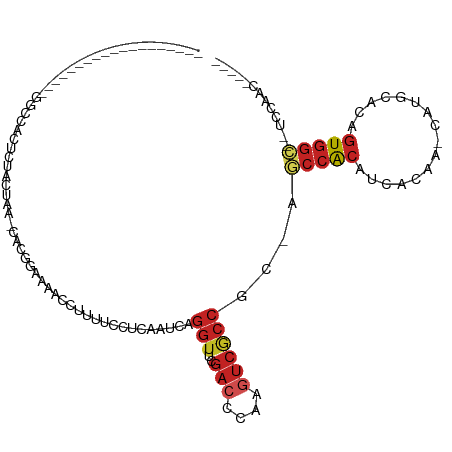
>dm3.chr2L 15650354 91 - 23011544 ------------------AUUCCACAUUACUUA-UACGGAAAACCUCUUCCUCAAUCAGGUUCGACCCAAGUCGCCGC-AGCCACAUCACAA-CAUGCACAGUGGC--UCCAAC----- ------------------...............-...((((......)))).......((..((((....))))))(.-((((((.......-........)))))--).)...----- ( -16.56, z-score = -2.09, R) >droSec1.super_3 1905058 82 - 7220098 ----------------------------ACUAACUACGGAAAACCUUUUCCUCAAUCAGGUUCGACCCAAGUCGCCGC-AGCCACAUCACAA-CAUGCACAGUGGC--UCCAAC----- ----------------------------.........(((((....))))).......((..((((....))))))(.-((((((.......-........)))))--).)...----- ( -17.06, z-score = -2.03, R) >droYak2.chr2L 2654299 91 + 22324452 ------------------UGGCUACUUUACUUA-UACGAAAAAUCUCGUCUUGAAUCAGGUUCGACCUAAGUCGCCGC-AGCCACAUCACAA-CAUGCACAGUGGC--UCCAAC----- ------------------.(((.((((...(((-.((((......))))..)))...(((.....))))))).)))(.-((((((.......-........)))))--).)...----- ( -20.46, z-score = -1.77, R) >droEre2.scaffold_4929 7395769 91 - 26641161 ------------------AGGCCACUUUACUUA-UGUGAAAUAUUUUGUCCCGCAUCAGGUUCGACCCAAGUCGCCGC-AGCCACAUCACAA-CAUGCACAGUGGC--UCCAAC----- ------------------.(((.(((((((...-.))))).....(((..((......))..))).....)).)))(.-((((((.......-........)))))--).)...----- ( -20.96, z-score = -1.16, R) >droAna3.scaffold_12916 15520855 86 + 16180835 ------------------GGGGUUCUCUGUUUAUCA------ACUCCGAUCUCAUACAGGUUCGACCCAAGUCACCGC-AACCACACCACAA-CAUGCACAGUGGC--UCAAAC----- ------------------.(((((.(((((..(((.------.....))).....)))))...))))).((((((.((-(............-..)))...)))))--).....----- ( -17.34, z-score = -0.83, R) >dp4.chr4_group4 4451227 87 - 6586962 ------------------UGGGCACUCCACUAAUCUUUGA-----UUUCCUUCCCUCAGGUUCGACCAAAGUCGCCGC-AGCCACAUCACAA-CAUGCACAGUGGC--UCCAAC----- ------------------..(((...............((-----...(((......))).))(((....))))))(.-((((((.......-........)))))--).)...----- ( -18.56, z-score = -0.90, R) >droPer1.super_93 73461 87 - 177177 ------------------UGGGCAUUCCACUAAUCUUUGA-----UUUCCUUUCCUCAGGUUCGACCAAAGUCGCCGC-AGCCACAUCACAA-CAUGCACAGUGGC--UCCAAC----- ------------------..(((...............((-----...(((......))).))(((....))))))(.-((((((.......-........)))))--).)...----- ( -18.56, z-score = -1.02, R) >droWil1.scaffold_180772 6058748 81 - 8906247 ----------------------CAUUCCACUGACGUUU-------UCAAUUUCACUCAGGUUCGACCCAAGUCACCGC-AACCACAUCAUAG-CUUGCACAGUGGU--UCCAAU----- ----------------------....((((((......-------.............(((..(((....))))))((-((.(........)-.)))).)))))).--......----- ( -16.80, z-score = -2.71, R) >anoGam1.chr3R 37573260 107 - 53272125 UCUUCUCUCUCUCUCGUUGCUUCACCCUACUCUUCUCU---CCCUUUCCCCCCCUGCAGGUGCGUCCACGUUCACCUC-AGCCACACCACGG----GCACGGUGGCAUUCCGGUC---- .............(((.(((..((((............---.............((.(((((((....))..))))))-)(((........)----))..)))))))...)))..---- ( -19.30, z-score = -0.02, R) >apiMel3.Group5 9631758 98 - 13386189 ---------------------CAACCCUCCCAGCCACCGGAAUAUUCAGCUACGAAUUCCGGUAAUGAAACUCACCGCAAAUCGAAACGUAAGUAUCCGAUAUAUCAAUCCAUUAUACU ---------------------...........((.((((((((..((......))))))))))..((.....))..))..((((..((....))...)))).................. ( -15.70, z-score = -1.68, R) >droSim1.chr2L 15403635 91 - 22036055 ------------------AUUCCACUUUACUAA-UACGGAAAACCUUUUCCUCAAUCAGGUUCGACCCAAGUCGCCGC-AGCCACAUCACAA-CAUGCACAGUGGC--UCCAAC----- ------------------...............-...(((((....))))).......((..((((....))))))(.-((((((.......-........)))))--).)...----- ( -17.06, z-score = -2.21, R) >consensus ___________________GGCCACUCUACUAA_CACGGAAAACCUUUUCCUCAAUCAGGUUCGACCCAAGUCGCCGC_AGCCACAUCACAA_CAUGCACAGUGGC__UCCAAC_____ ..........................................................(((..(((....))))))....(((((................)))))............. ( -5.88 = -6.58 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:23 2011