
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,635,202 – 15,635,304 |
| Length | 102 |
| Max. P | 0.745530 |

| Location | 15,635,202 – 15,635,304 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.42 |
| Shannon entropy | 0.47983 |
| G+C content | 0.39614 |
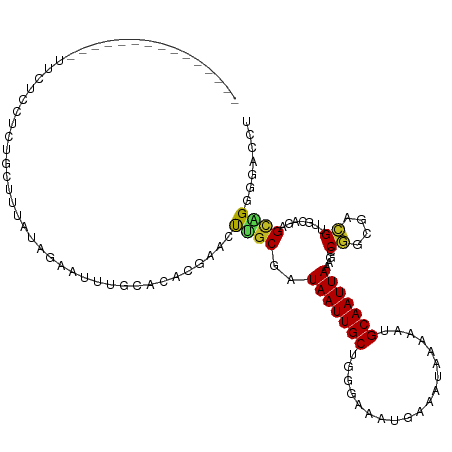
| Mean single sequence MFE | -27.24 |
| Consensus MFE | -6.86 |
| Energy contribution | -6.10 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
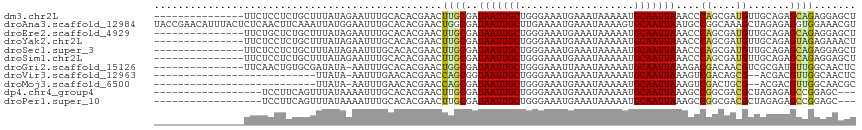
>dm3.chr2L 15635202 102 + 23011544 ---------------UUCUCCUCUGCUUUAUAGAAUUUGCACACGAACUUGCGAUAAUUGCUGGGAAAUGAAAUAAAAAUGCAAUUAAACCCAGCGAUGUUGCAGAGCAGAGGAGCU ---------------..((((((((((((......((((....))))...((((((.((((((((........................)))))))))))))))))))))))))).. ( -37.76, z-score = -5.68, R) >droAna3.scaffold_12984 442637 117 - 754457 UACCGAACAUUUACUCUCAACUUCAAAUUAUGGAAUUUGCACACGAACUGGCGAUAAUUGCUUGAAAAUGAAAUAAAAGUGCAAUUAAUGCCGGCAAAGCUAGAGAGGUGGAAACGU .............(((((.....((((((....))))))........((((((.(((((((...................))))))).))))))........)))))..(....).. ( -26.31, z-score = -1.98, R) >droEre2.scaffold_4929 7378427 102 + 26641161 ---------------UUCUGCUCUGCUUUAUAGAAUUUGCACACGAACUUGCGAUAAUUGCUGGGAAAUGAAAUAAAAAUGCAAUUAAACCCAGCGAUGUUGCAGAGCAGAGGAGCU ---------------(((((((((((..........(((((........)))))..(((((((((........................)))))))))...)))))))))))..... ( -34.86, z-score = -4.50, R) >droYak2.chr2L 2639064 102 - 22324452 ---------------UUCUCCUCUGCUUUAUAGAAUUUGCACACGAACUUGCGAUAAUUGCUGGGAAAUGAAAUAAAAAUGCAAUUAAACCCAGCGAUGUUGCAGAGUAGAGAAACU ---------------(((((((((((..........(((((........)))))..(((((((((........................)))))))))...))))))..)))))... ( -29.26, z-score = -4.20, R) >droSec1.super_3 1889728 102 + 7220098 ---------------UUCUCCUCUGCUUUAUAGAAUUUGCACACGAACUUGCGAUAAUUGCUGGGAAAUGAAAUAAAAAUGCAAUUAAACCCAGCGAUGUUGCAGAGCAGAGGAGCU ---------------..((((((((((((......((((....))))...((((((.((((((((........................)))))))))))))))))))))))))).. ( -37.76, z-score = -5.68, R) >droSim1.chr2L 15387685 102 + 22036055 ---------------UUCUCCUCUGCUUUAUAGAAUUUGCACACGAACUUGCGAUAAUUGCUGGGAAAUGAAAUAAAAAUGCAAUUAAACCCAGCGAUGUUGCAGAGCAGAGGAGCU ---------------..((((((((((((......((((....))))...((((((.((((((((........................)))))))))))))))))))))))))).. ( -37.76, z-score = -5.68, R) >droGri2.scaffold_15126 2546113 101 - 8399593 ---------------UUCAACUGUGCGAUAUA-AAUUUGCACACGAACUGGCGAUAAUUGCUGGGAAAUUAAAUAAAAAUGCAAUUAAAGACGACAACGUCGCGAUGUUGGCAACUC ---------------.((((((((((((....-...))))))).......((..(((((((...................)))))))..((((....))))))...)))))...... ( -23.31, z-score = -2.05, R) >droVir3.scaffold_12963 7843671 87 - 20206255 ---------------------------UUAUA-AAUUUGAACACGAACCAGCGGUAAUUGCUGGGAAAUGAAAUAAAAAUGCAAUUAAAGUCGACAGCG--ACGACGUUGGCAACUC ---------------------------.....-..............((((((.(((((((...................)))))))..((((....))--))..))))))...... ( -18.61, z-score = -1.75, R) >droMoj3.scaffold_6500 11273924 87 - 32352404 ---------------------------UUAUA-AAUUUGAACACGAACCAGCGAUAAUUGCUGGGAAAUGAAAUAAAAAUGCAAUUAAAGUCGACUGCG--ACGACGUUGGCAACGC ---------------------------.....-..........((..((((((.(((((((...................)))))))..((((....))--))..))))))...)). ( -18.61, z-score = -1.95, R) >dp4.chr4_group4 4432552 96 - 6586962 ------------------UCCUUCAGUUUAUAAAAUUUGCACACGAACUUGCGAUAAUUGCUGGGAAAUGAAAUAAAAAUGCAAUUAAAGCCGGCGACGCUAGAGAGCCGGAGC--- ------------------((((.(((((........(((((........))))).)))))..))))........................(((((...........)))))...--- ( -17.70, z-score = 0.45, R) >droPer1.super_10 2269427 96 - 3432795 ------------------UCCUUCAGUUUAUAAAAUUUGCACACGAACUUGCGAUAAUUGCUGGGAAAUGAAAUAAAAAUGCAAUUAAAGCCGGCGACGCUAGAGAGCCGGAGC--- ------------------((((.(((((........(((((........))))).)))))..))))........................(((((...........)))))...--- ( -17.70, z-score = 0.45, R) >consensus _______________UUCUCCUCUGCUUUAUAGAAUUUGCACACGAACUUGCGAUAAUUGCUGGGAAAUGAAAUAAAAAUGCAAUUAAAGCCGGCGACGUUGCAGAGCAGGGGACCU ................................................((((..(((((((...................)))))))....((....)).......))))....... ( -6.86 = -6.10 + -0.76)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:19 2011