
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,619,169 – 15,619,272 |
| Length | 103 |
| Max. P | 0.500000 |

| Location | 15,619,169 – 15,619,272 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Shannon entropy | 0.14571 |
| G+C content | 0.45379 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -22.01 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 15619169 103 + 23011544 UGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAAAA-GCCAAAAACCCAC---------------- .(((.....(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))...)-))....)))....---------------- ( -31.40, z-score = -2.45, R) >droSim1.chr2L 15371586 103 + 22036055 UGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAAAA-GCCAAAAACCCAC---------------- .(((.....(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))...)-))....)))....---------------- ( -31.40, z-score = -2.45, R) >droSec1.super_3 1873994 103 + 7220098 UGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGUCUUUACGAGCCAAGGCAAAA-GCCAAAAACCCAC---------------- .(((.....(((..(((((.(((((((((((((....))))))......(((((........)))))......))))))).)))))...)-))....)))....---------------- ( -29.80, z-score = -1.89, R) >droYak2.chr2L 2613864 103 - 22324452 UGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAAAA-GCCAAAAACCCAC---------------- .(((.....(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))...)-))....)))....---------------- ( -31.40, z-score = -2.45, R) >droEre2.scaffold_4929 7362378 103 + 26641161 UGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAAAA-GCCAAAAACCCAC---------------- .(((.....(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))...)-))....)))....---------------- ( -31.40, z-score = -2.45, R) >droAna3.scaffold_12984 408915 102 - 754457 UGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAAAA-GCCAAAA-CCCAC---------------- .(((.....(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))...)-))...))-)....---------------- ( -31.00, z-score = -2.34, R) >dp4.chr4_group4 4409457 103 - 6586962 UGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAAAA-GCCGAAUUCCCAC---------------- .......(.(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))...)-)).).........---------------- ( -31.00, z-score = -2.34, R) >droPer1.super_10 2246553 103 - 3432795 UGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAAAA-GCCGAAUUCCCAC---------------- .......(.(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))...)-)).).........---------------- ( -31.00, z-score = -2.34, R) >droWil1.scaffold_180764 2594977 120 - 3949147 UGUUGCUCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAAAAAGCCAAAAAACCCCACAAUCCCCACUAACC .(((.....(((..(((((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))))....))).....)))................... ( -31.10, z-score = -2.08, R) >droMoj3.scaffold_6500 21056706 101 + 32352404 UGUUGCUGUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAG-CAAAA-GCCAAAACCCAG----------------- .(((..((.(((....(((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).)))-....)-)))).)))....----------------- ( -26.70, z-score = -1.18, R) >droVir3.scaffold_12963 12342961 101 + 20206255 UGUUGCUGUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGGCAAG-CAAAA-GCCAAAACCCAG----------------- ....(((.((((..(((((...........(((...)))((((((((..(((((........))))).))))))))))))).))-))..)-))..........----------------- ( -28.70, z-score = -1.35, R) >droGri2.scaffold_15126 5837128 100 + 8399593 UGUUGCUGUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAG-CAAAA-GA-AAAACCCAG----------------- ..(((((..(((.(................(((...)))((((((((..(((((........))))).))))))))))))..))-)))..-..-.........----------------- ( -24.10, z-score = -0.69, R) >consensus UGUUGCCCUGCUACGCCUUUGUUUGUAUUGCCCUCAGGGGUAAAGUUUUUAGGAUAAUAACUUCCUGCGACUUUACGAGCCAAGGCAAAA_GCCAAAAACCCAC________________ .((.((...)).))(.(((.(((((((((((((....))))))((((..(((((........))))).)))).))))))).))).).................................. (-22.01 = -22.18 + 0.17)
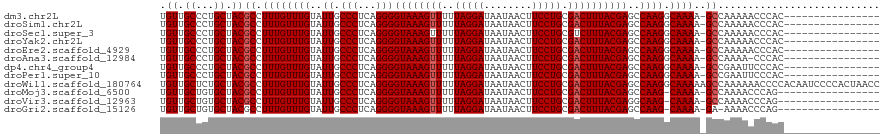

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:18 2011