
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,604,788 – 15,604,884 |
| Length | 96 |
| Max. P | 0.770734 |

| Location | 15,604,788 – 15,604,884 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.60 |
| Shannon entropy | 0.66369 |
| G+C content | 0.45853 |
| Mean single sequence MFE | -30.81 |
| Consensus MFE | -10.66 |
| Energy contribution | -10.11 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
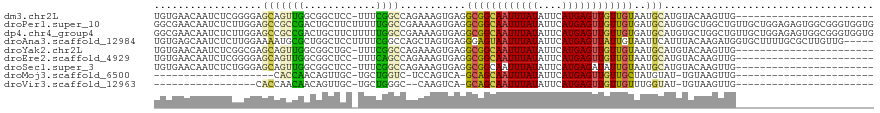
>dm3.chr2L 15604788 96 - 23011544 UGUGAACAAUCUCGGGGAGCAGUUGGCGGCUCC-UUUCGGCCAGAAAGUGAGGCGGCAAUUUAUAUUCAUGAGUUGUUGUAAUGCAUGUACAAGUUG----------------------- ....(((((.((((.(((..(((((.((.((((-((((.....))))).))).)).)))))....))).)))))))))...................----------------------- ( -27.90, z-score = -1.09, R) >droPer1.super_10 2226415 120 + 3432795 GGCGAACAAUCUCUUGGAGCCGCCGACUGCUUCUUUUUGGCCGAAAAGUGAGGCGGCAAUUUAUAUUCAUGAGUUGUUGUGAUGCAUGUGCUGGCUGUUGCUGGAGAGUGGCGGGUGGUG (((...(((....)))..)))((((.(((((.(((((..((.....(((.(.((((((((((((....))))))))))))...((....))).)))...))..))))).))))).)))). ( -42.00, z-score = -1.38, R) >dp4.chr4_group4 4388853 120 + 6586962 GGCGAACAAUCUCUUGGAGCCGCCGACUGCUUCUUUUUGGCCGAAAAGUGAGGCGGCAAUUUAUAUUCAUGAGUUGUUGUGAUGCAUGUGCUGGCUGUUGCUGGAGAGUGGCGGGUGGUG (((...(((....)))..)))((((.(((((.(((((..((.....(((.(.((((((((((((....))))))))))))...((....))).)))...))..))))).))))).)))). ( -42.00, z-score = -1.38, R) >droAna3.scaffold_12984 389804 115 + 754457 UGUGAGCAAUCUCUUGGAAAAUGCGCUGGCUCCUUUUCGGCCAGCUAGUGAGGGAGUAAUUUAUAUUCAUGAGUUAUUGUAAUUCAUUUACAAGAUGGUGCUUUUGCGCUUGUUG----- .(..(((.((((((((........(((((((.......))))))).(((((.(.((((((((((....)))))))))).)...)))))..))))).)))((....)))))..)..----- ( -33.20, z-score = -1.60, R) >droYak2.chr2L 2598368 96 + 22324452 UGUGAACAAUCUCGGCGAGCAGUUGGCGGCUGC-UUUCGGCCAGAAAGUGAGGCGGCAAUUUAUAUUCAUGAGUUGUUGUAAUGCAUGUACAAGUUG----------------------- ..((.(((.(((.((((((((((.....)))))-))...))))))..(((..((((((((((((....))))))))))))..))).))).)).....----------------------- ( -29.10, z-score = -1.03, R) >droEre2.scaffold_4929 7346120 96 - 26641161 UGUGAACAAUCUCGGGGAGCAGUUGGCGGCUCC-UUUCAGCCAGAAAGUGAGGCGGCAAUUUAUAUUCAUGAGUUGUUGUAAUGCAUGUACAAGUUG----------------------- ....(((((.((((.(((..(((((.((.((((-((((.....))))).))).)).)))))....))).)))))))))...................----------------------- ( -27.20, z-score = -0.98, R) >droSec1.super_3 1859434 96 - 7220098 UGUGAACAAUCUCUGGGAGCAGUUGGCGGCUCC-UUUCGGCCAGAAAGUGAGGCGGCAAUUUAUAUUCAUGAGAUAUUGUAAUGCAUGUACAAGUUG----------------------- ....(((.(((((((((...(((((.((.((((-((((.....))))).))).)).)))))....)))).))))).(((((.......)))))))).----------------------- ( -27.80, z-score = -1.29, R) >droMoj3.scaffold_6500 21036771 73 - 32352404 --------------------CACCAACAGUUGC-UGCUGGUC-UCCAGUCA-GCAGCAAUUUAUAUUCAUGAGUUGUUGCUAUGUAU-UGUAAGUUG----------------------- --------------------........(((((-((((((..-.)))).))-)))))((((((((..(((.(((....))))))...-)))))))).----------------------- ( -23.00, z-score = -2.87, R) >droVir3.scaffold_12963 12309698 75 - 20206255 -----------------CACCAACAACAGUUGC-UGCUGGGC--CAAGUCA-GCAGCAAUUUAUAUUCAUGAGUUGUUGUUUGGUAU-UGUAAGUUG----------------------- -----------------.(((((((((((((((-((((((..--....)))-))))))))((((....))))..))))).)))))..-.........----------------------- ( -25.10, z-score = -2.83, R) >consensus UGUGAACAAUCUCUGGGAGCAGUCGACGGCUCC_UUUCGGCCAGAAAGUGAGGCGGCAAUUUAUAUUCAUGAGUUGUUGUAAUGCAUGUACAAGUUG_______________________ ..................(((((((............))))...........((((((((((((....))))))))))))..)))................................... (-10.66 = -10.11 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:17 2011