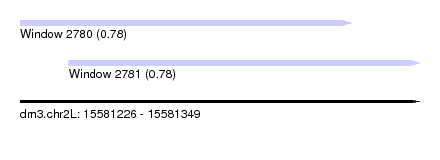
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,581,226 – 15,581,349 |
| Length | 123 |
| Max. P | 0.784086 |

| Location | 15,581,226 – 15,581,328 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 64.59 |
| Shannon entropy | 0.72803 |
| G+C content | 0.59352 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -10.23 |
| Energy contribution | -10.73 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.784086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15581226 102 + 23011544 UUUGAAAAACCCCACGCCCA------UUGCCCACCAGCCAUCCGCCUCCAUCUCCGGCGACUAUGAGAACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCC---- ...............((...------((((............((((.........))))..........(((((((.(....))))))))......))))))......---- ( -22.30, z-score = -0.85, R) >droSim1.chr2L 15339217 102 + 22036055 UUCGAAAAACCCCACGCCCA------UUGCCCACCAGCCAUCCACCUCCAUCUCCGGCGACUAUGAGAACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCC---- ...............((...------((((....................((((.(....)...)))).(((((((.(....))))))))......))))))......---- ( -19.90, z-score = -0.45, R) >droSec1.super_3 1842862 102 + 7220098 UUCGAAAAACCCCACGCCCA------UUGCCCACCAGCCAUCCGCCUCCAUCUCCGGCGACUAUGAGAACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCC---- ...............((...------((((............((((.........))))..........(((((((.(....))))))))......))))))......---- ( -22.30, z-score = -0.67, R) >droYak2.chr2L 2581936 108 - 22324452 UCCGAAAAACCCCACGUCCACGCCCGUUGCCCAACAGCCAUCCGCAUCCAUCUCCGGCGACUAUGAGAACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCC---- .....................((..((((.....))))....(((((...((((.(....)...)))).(((((((.(....))))))))....))))).))......---- ( -25.10, z-score = -0.58, R) >droEre2.scaffold_4929 7329688 106 + 26641161 UCCGAAAAACCCCACGCCCA------UUGCCCAACAGUCAUCCGCCUCCAUCUCCGGCGACUAUGAGAACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCCAUCC ...............((...------((((.......((((.((((.........))))...))))...(((((((.(....))))))))......)))))).......... ( -23.60, z-score = -1.00, R) >droAna3.scaffold_12984 275592 92 - 754457 UCCGCAACACUCCUC-----------UGGCCU--CAGCCGCGC---UCUCUUUCCCACAAAUGGAAAAGCGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACCUCC---- ..((((.........-----------.(((..--..)))((((---(...(((((.......))))))))))((((((....).)))))......)))).........---- ( -27.90, z-score = -1.90, R) >dp4.chr4_group4 4370200 90 - 6586962 ------------AGCACUCA------CUCCGCAUCCCCCACCCGGAUCUCCCUCCUCCAUCUU---ACACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGGGCUGGCCCC- ------------........------...(((((.........(((......)))........---...(((((((.(....))))))))....))))).(((....))).- ( -24.90, z-score = -1.21, R) >droPer1.super_10 2207450 90 - 3432795 ------------AGCACUCA------CUCCGCAUCCCCCACCCGGAUCUCCCUCCUCCAUCUU---ACACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGGGCUGGCCCC- ------------........------...(((((.........(((......)))........---...(((((((.(....))))))))....))))).(((....))).- ( -24.90, z-score = -1.21, R) >droVir3.scaffold_12963 12288003 101 + 20206255 CACCAACGACUCCCCCCUCAG------CGCACCUCACCCCUUU-CACACCUCUUCCUCACCAGCGUAAAGGCGGUUGCUAAUGAUGCGCGAAAUAUGCGAUGGCUGCC---- ..................(((------((((....((((((((-.((.(.............).))))))).))))))........((((.....))))...))))..---- ( -16.82, z-score = 1.20, R) >droMoj3.scaffold_6500 21013021 89 + 32352404 ---------CUGCAUCCACC-------CAUACACCACCCCCUC-UCCCCACACCCAUCAUCCUUGCGAGGACGCGUGCUAAUGACGCGCAAAAUAUGCGUUGCCUC------ ---------..(((...((.-------((((........((((-......................))))..((((((....).)))))....)))).)))))...------ ( -14.95, z-score = -1.01, R) >consensus U_CGAAAAACCCCACGCCCA______UUGCCCACCAGCCAUCCGCCUCCAUCUCCGGCAACUAUGAGAACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCC____ .....................................................................(((((((.(....))))))))...................... (-10.23 = -10.73 + 0.50)
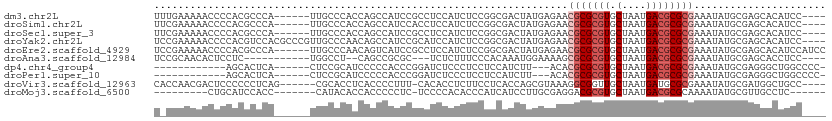

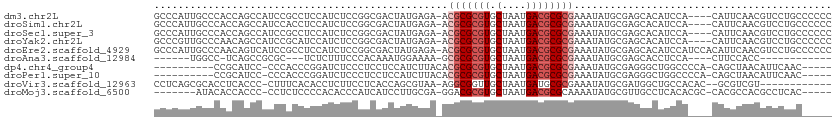
| Location | 15,581,241 – 15,581,349 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 64.10 |
| Shannon entropy | 0.74550 |
| G+C content | 0.59843 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -10.23 |
| Energy contribution | -10.73 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.779213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15581241 108 + 23011544 GCCCAUUGCCCACCAGCCAUCCGCCUCCAUCUCCGGCGACUAUGAGA-ACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCCA----CAUUCAACGUCCUGCCCCCC ((...((((............((((.........)))).........-.(((((((.(....))))))))......)))))).......----.................... ( -22.00, z-score = -0.10, R) >droSim1.chr2L 15339232 108 + 22036055 GCCCAUUGCCCACCAGCCAUCCACCUCCAUCUCCGGCGACUAUGAGA-ACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCCA----CAUUCAACGUCCUGCCCCCC ..................................((((((..((((.-.(((((((.(....)))))))).....((.((.....))))----..))))..)))..))).... ( -20.60, z-score = -0.15, R) >droSec1.super_3 1842877 108 + 7220098 GCCCAUUGCCCACCAGCCAUCCGCCUCCAUCUCCGGCGACUAUGAGA-ACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCCA----CAUUCAACGUCCUGCCCCCC ((...((((............((((.........)))).........-.(((((((.(....))))))))......)))))).......----.................... ( -22.00, z-score = -0.10, R) >droYak2.chr2L 2581957 108 - 22324452 GCCCGUUGCCCAACAGCCAUCCGCAUCCAUCUCCGGCGACUAUGAGA-ACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCCA----CAUUCAACGUCCUGCCCCCC (..(((((............((((((...((((.(....)...))))-.(((((((.(....))))))))....))))).)........----....)))))..)........ ( -24.95, z-score = -0.45, R) >droEre2.scaffold_4929 7329703 112 + 26641161 GCCCAUUGCCCAACAGUCAUCCGCCUCCAUCUCCGGCGACUAUGAGA-ACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCCAUCCACAUUCAACGUCCUGCCCCCC ((...((((.......((((.((((.........))))...))))..-.(((((((.(....))))))))......))))))............................... ( -23.30, z-score = -0.31, R) >droAna3.scaffold_12984 275607 86 - 754457 ------UGGCC-UCAGCCGCGC---UCUCUUUCCCACAAAUGGAAAA-GCGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACCUCCA----CUUCCACC------------ ------.(((.-...)))..((---((.((((.(((....))).)))-)(((((((.(....))))))))........)))).......----........------------ ( -26.30, z-score = -1.92, R) >dp4.chr4_group4 4370210 96 - 6586962 ----------CCGCAUCC-CCCACCCGGAUCUCCCUCCUCCAUCUUACACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGGGCUGGCCCCA-CAGCUAACAUUCAAC----- ----------.(((((..-.......(((......)))...........(((((((.(....))))))))....)))))..(((((......-)))))..........----- ( -27.80, z-score = -2.42, R) >droPer1.super_10 2207460 96 - 3432795 ----------CCGCAUCC-CCCACCCGGAUCUCCCUCCUCCAUCUUACACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGGGCUGGCCCCA-CAGCUAACAUUCAAC----- ----------.(((((..-.......(((......)))...........(((((((.(....))))))))....)))))..(((((......-)))))..........----- ( -27.80, z-score = -2.42, R) >droVir3.scaffold_12963 12288018 97 + 20206255 CCUCAGCGCACCUCACCC-CUUUCACACCUCUUCCUCACCAGCGUAA-AGGCGGUUGCUAAUGAUGCGCGAAAUAUGCGAUGGCUGCCACAC--GCGUCGU------------ .......(((....((((-((((.((.(.............).))))-))).))))))..((((((((.(......((....)).....).)--)))))))------------ ( -19.92, z-score = 1.43, R) >droMoj3.scaffold_6500 21013033 98 + 32352404 -------AUACACCACCC-CCUCUCCCCACACCCAUCAUCCUUGCGA-GGACGCGUGCUAAUGACGCGCAAAAUAUGCGUUGCCUCACACGC-CACGCCACGCCUCAC----- -------...........-..........................((-((.(((((((....).))))).......((((.((.......))-.))))...)))))..----- ( -20.60, z-score = -2.12, R) >consensus GCCCAUUGCCCACCAGCCACCCGCCUCCAUCUCCGGCGACUAUGAGA_ACGCGCGUGCUAAUGACGCGCGAAAUAUGCGAGCACAUCCA____CAUUCAACGUCCUGC_____ .................................................(((((((.(....))))))))........................................... (-10.23 = -10.73 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:16 2011