| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,556,199 – 15,556,274 |
| Length | 75 |
| Max. P | 0.997131 |

| Location | 15,556,199 – 15,556,274 |
|---|---|
| Length | 75 |
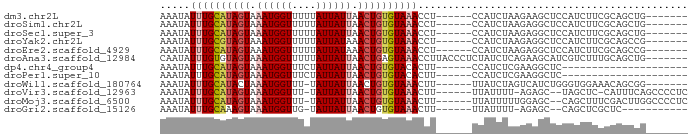
| Sequences | 12 |
| Columns | 88 |
| Reading direction | forward |
| Mean pairwise identity | 73.49 |
| Shannon entropy | 0.53602 |
| G+C content | 0.33321 |
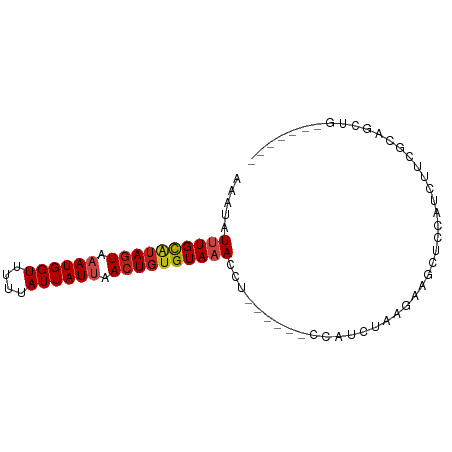
| Mean single sequence MFE | -14.58 |
| Consensus MFE | -10.46 |
| Energy contribution | -10.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
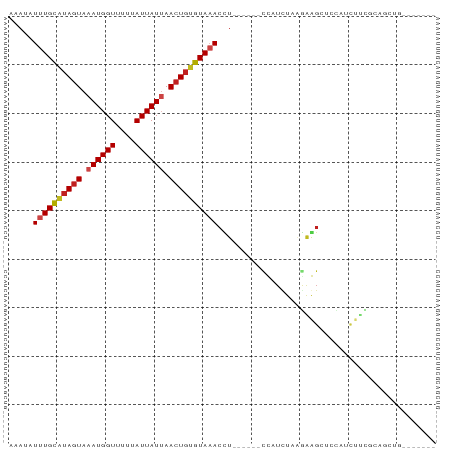
| Mean z-score | -2.11 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15556199 75 + 23011544 AAAUAUUUGCAUAGUAAAUGGUUUUUAUUAUUAACUGUGUAAACCU------CCAUCUAAGAAGCUCCAUCUUCGCAGCUG------- .....((((((((((.((((((....)))))).))))))))))...------..........((((.(......).)))).------- ( -15.90, z-score = -3.03, R) >droSim1.chr2L 15314232 75 + 22036055 AAAUAUUUGCAUAGUAAAUGGUUUUUAUUAUUAACUGUGUAAACCU------CCAUCUAAGAGGCUCCAUCUUCGCAGCUG------- .....((((((((((.((((((....)))))).))))))))))(((------(.......)))).................------- ( -16.60, z-score = -2.67, R) >droSec1.super_3 1818028 75 + 7220098 AAAUAUUUGCAUAGUAAAUGGUUUUUAUUAUUAACUGUGUAAACCU------CCAUCUAAGAGGCUCCAUCUUCGCAGCUG------- .....((((((((((.((((((....)))))).))))))))))(((------(.......)))).................------- ( -16.60, z-score = -2.67, R) >droYak2.chr2L 2555379 75 - 22324452 AAAUAUUUGCGUAGUAAAUGGUUUUUAUUAUAAACUGUGUAAACCU------CCAUCUAAGAGGCUCCAUCUUCGCAGCCG------- .....((((((((((..(((((....)))))..))))))))))...------..........((((.(......).)))).------- ( -13.10, z-score = -1.24, R) >droEre2.scaffold_4929 7304976 75 + 26641161 AAAUAUUUGCAUAGUAAAUGGUUUUUAUUAUAAACUGUGUAAACCU------CCAUCUAAGAGGCUCCAUCUUCGCAGCCG------- .....((((((((((..(((((....)))))..))))))))))...------..........((((.(......).)))).------- ( -16.50, z-score = -2.85, R) >droAna3.scaffold_12984 235339 81 - 754457 CAAUAUUUGUGUAGUAAAUGGUUUUUAUUAUUAACUGAGUAAACCUUACCCUCUAUCUCAGAAGCAUCGUCUUUGCAGCUG------- .....((((..((((.((((((....)))))).))))..))))...............(((..(((.......)))..)))------- ( -10.50, z-score = 0.20, R) >dp4.chr4_group4 4342092 61 - 6586962 AAAUAUUUGCAUAGUAAAUGGUUUCUAUUAUUAACUGUGUACACUU------CCAUCUCGAAGGCUC--------------------- .......((((((((.((((((....)))))).))))))))..(((------(......))))....--------------------- ( -13.60, z-score = -3.12, R) >droPer1.super_10 2179182 61 - 3432795 AAAUAUUUGCAUAGUAAAUGGUUUCUAUUAUUAACUGUGUACACUU------CCAUCUCGAAGGCUC--------------------- .......((((((((.((((((....)))))).))))))))..(((------(......))))....--------------------- ( -13.60, z-score = -3.12, R) >droWil1.scaffold_180764 2508155 74 - 3949147 AAAUAUUUGCAUACUAAAUGGUUU-UAUUAUUAACUGUGUAAACUU------UUAUCUAGUCAUCUGGGUGGAAACAGCGG------- .....((((((((.(.((((((..-.)))))).).)))))))).((------((((((((....)))))))))).......------- ( -13.50, z-score = -1.49, R) >droVir3.scaffold_12963 12258822 77 + 20206255 AAAUAUUUGCAUAGUAAAUGGUUU-UAUUAUUAACUGUGUAAACUU------UUAUUUU-AGAGC--UAGCUC-CAUUUCAGCCCCUC (((..((((((((((.((((((..-.)))))).))))))))))..)------)).....-.(((.--...)))-.............. ( -14.00, z-score = -1.72, R) >droMoj3.scaffold_6500 20980412 79 + 32352404 AAAUAUUUGCAUAGUAAAUGGUUU-UAUUAUUAACUGUGUAAACUU------UUAUUUUUGGAGC--CAGCUUUCGACUUGGCCCCUC (((..((((((((((.((((((..-.)))))).))))))))))..)------))......((.((--(((........))))).)).. ( -19.90, z-score = -3.16, R) >droGri2.scaffold_15126 5771296 67 + 8399593 AAAUAUUUGCAAAGUAAAUGGUUG-UAUUAUUAACUGUGUAAACUU------UUAUUUU-AGAGC--CAGCUCGCUC----------- (((..((((((.(((.((((((..-.)))))).))).))))))..)------)).....-.((((--......))))----------- ( -11.20, z-score = -0.52, R) >consensus AAAUAUUUGCAUAGUAAAUGGUUUUUAUUAUUAACUGUGUAAACCU______CCAUCUAAGAAGCUCCAUCUUCGCAGCUG_______ .....((((((((((.((((((....)))))).))))))))))............................................. (-10.46 = -10.90 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:14 2011