
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,432,594 – 1,432,705 |
| Length | 111 |
| Max. P | 0.508995 |

| Location | 1,432,594 – 1,432,705 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.69 |
| Shannon entropy | 0.46356 |
| G+C content | 0.52952 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -16.13 |
| Energy contribution | -16.39 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1432594 111 - 23011544 -CAGUGGGU-CCGGUAUCAAUACGCGCCUAAUGAGUCAAGUGGCCG-AAAGGAUUUUCGGUGGGUGCGGAUGCCGAUGCGGUUGUAUCUCUGUAUCUCAGUAUCUCACUGUCUG------ -(((((((.-..((((((....((((((((.((((........((.-...))...)))).))))))))))))))(((((((........))))))).......)))))))....------ ( -43.10, z-score = -3.24, R) >droPer1.super_8 1167725 118 - 3966273 CCAGGCAGC-UUCGUAUCAAUACGCGCCUAAUGAGUCAAGUGGCCAAAAAGGAUUUUCGGUGGGUGCGGACUGCGGUCUGG-UGUAUCUCUGUAUCUCAGUAUCUCUCUGGCUGCUGUCU ...((((((-(..(((......((((((((..(((((...(......)...)))))....))))))))...)))((((((.-.((((....))))..))).))).....))))))).... ( -33.20, z-score = 0.16, R) >dp4.chr4_group3 9960478 118 - 11692001 CCAGGCAGC-UUCGUAUCAAUACGCGCCUAAUGAGUCAAGUGGCCAAAAAGGAUUUUCGGUGGGUGCGGACUGCGGUCUGG-UGUAUCUCUGUAUCUCAGUAUCUCUCUGGCUGCUGUCU ...((((((-(..(((......((((((((..(((((...(......)...)))))....))))))))...)))((((((.-.((((....))))..))).))).....))))))).... ( -33.20, z-score = 0.16, R) >droAna3.scaffold_12916 4895213 106 - 16180835 -CAGUCGGC-CUGGUAUCAAUACCCGCCUAAUGAGUCAAGUGGCCGAAAAGGAUUUUCGGUGGGUGCGGA-UGCGGCUU----GUAUCUGU-UCCCUGUCUGUCUCGCUGUCUG------ -(((.((((-..((((....))))........(((.((....(((((((.....)))))))(((.(((((-(((.....----))))))))-.)))....)).))))))).)))------ ( -38.30, z-score = -2.38, R) >droEre2.scaffold_4929 1478130 110 - 26641161 -CAGUGGGU-CCGGUAUCAAUACGCGCCUAAUGAGUCAAGUGGCCG-AAAGGA-UUUCGGUGGGUGCGGAUGCCGAUGCGGUUGUAUCUCUGUAUCUCCGUAUCUCACUGUCUG------ -(((((((.-.(((((((....((((((((.((((........((.-...)).-.)))).)))))))))))))))((((((..((((....))))..)))))))))))))....------ ( -44.60, z-score = -3.49, R) >droYak2.chr2L 1407706 112 - 22324452 -CAGUGGGUCCCGGUAUCAAUACGCGCCUAAUGAGUCAAGUGGCCG-AAAGGAUUUUCGGUGGGUGCGGAUGUCAAUGCGGUUGUAUCUUUGUAUCUCGGUAUCUCACUGUCUG------ -(((((((..(((..(((((..((((((((.((((........((.-...))...)))).))))))))((((.((((...)))))))).))).))..)))...)))))))....------ ( -37.20, z-score = -1.92, R) >droSec1.super_14 1380145 111 - 2068291 -CAGUGGGU-CCGGUAUCAAUGCGCGCCUAAUGAGUCAAGUGGCCG-AAAGGAUUUUCGGUGGGUGCGGAUGCCGAUGCGGUUGUAUCUCUGUAUCUCCGUAUCUCACUGUCUG------ -(((((((.-.(((((((....((((((((.((((........((.-...))...)))).)))))))))))))))((((((..((((....))))..)))))))))))))....------ ( -45.00, z-score = -3.41, R) >droSim1.chr2L 1405738 111 - 22036055 -CAGUGGGU-CCGGUAUCAAUGCGCGCCUAAUGAGUCAAGUGGCCG-AAAGGAUUUUCGGUGGGUGCGGAUGCCGAUGCGGUUGUAUCUCUGUAUCUCCGUAUCUCACUGUCUG------ -(((((((.-.(((((((....((((((((.((((........((.-...))...)))).)))))))))))))))((((((..((((....))))..)))))))))))))....------ ( -45.00, z-score = -3.41, R) >droGri2.scaffold_15252 10543947 84 - 17193109 ----UGGCAUGGCGUAUCAAUGCUCGCCUAAUGAGUCAAGUGGCC--AAAGGAUUUU--GUGGGUGCG----UUUGUCUGGACGUCGCAACGUGGG------------------------ ----.((((.((((((((........(((..((.(((....))))--).))).....--...))))))----))))))...((((....))))...------------------------ ( -20.39, z-score = 1.51, R) >consensus _CAGUGGGU_CCGGUAUCAAUACGCGCCUAAUGAGUCAAGUGGCCG_AAAGGAUUUUCGGUGGGUGCGGAUGCCGAUGCGGUUGUAUCUCUGUAUCUCAGUAUCUCACUGUCUG______ .((((.((..............((((((((.((((........((.....))...)))).))))))))......(((.(((........))).))).......)).)))).......... (-16.13 = -16.39 + 0.26)
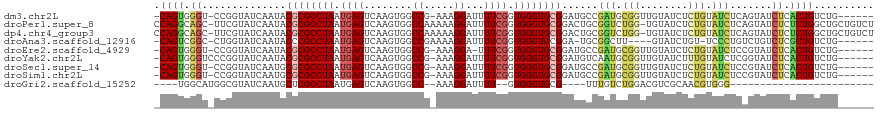
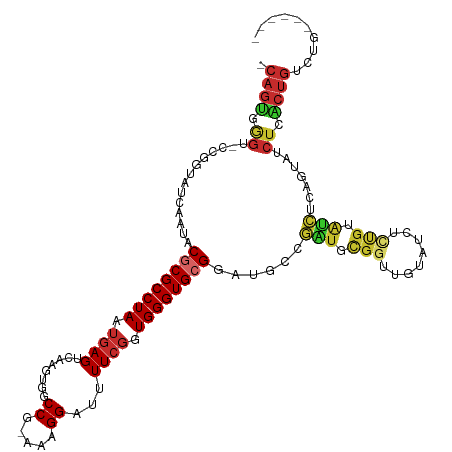
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:34 2011