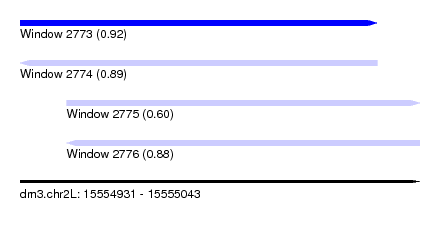
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,554,931 – 15,555,043 |
| Length | 112 |
| Max. P | 0.920308 |

| Location | 15,554,931 – 15,555,031 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 74.71 |
| Shannon entropy | 0.53527 |
| G+C content | 0.39681 |
| Mean single sequence MFE | -16.79 |
| Consensus MFE | -8.90 |
| Energy contribution | -9.01 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
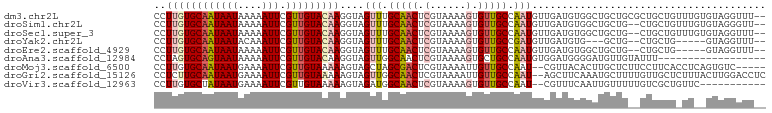
>dm3.chr2L 15554931 100 + 23011544 --AAACCUACACAAACAGCAGCGCAGCAGCCACAUCAACAUUGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUUUAUUAUUGCACAAGG --....................(((((.((((.........))))....(......).)))))......((((((.(((..(.......)..))).)))))) ( -17.70, z-score = -1.46, R) >droSim1.chr2L 15312987 98 + 22036055 --AACCCUACACAAACAGCAG--CAGCAGCCACAUCAACAUUGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUUUAUUAUUGCACAAGG --..................(--((((.((((.........))))....(......).)))))......((((((.(((..(.......)..))).)))))) ( -17.60, z-score = -1.92, R) >droSec1.super_3 1816783 98 + 7220098 --AAACCUACACAAACAGCAG--CAGCAGCCACAUCAACAUUGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUUUAUUAUUGCACAAGG --..................(--((((.((((.........))))....(......).)))))......((((((.(((..(.......)..))).)))))) ( -17.60, z-score = -1.92, R) >droYak2.chr2L 2554177 90 - 22324452 --AAACCUAC-----CAGCAG--CAGC---CACAUCAACAUCGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUGUAUUAUUGCACAAGG --........-----..((((--(.((---(...........)))....(......).)))))......((((((.(((..(.......)..))).)))))) ( -16.60, z-score = -1.51, R) >droEre2.scaffold_4929 7303679 93 + 26641161 --AAACCUAC-----CAGCAG--CAGCAGCCACAUCAACAUUGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUUUAUUAUUGCACAAGG --........-----.....(--((((.((((.........))))....(......).)))))......((((((.(((..(.......)..))).)))))) ( -17.60, z-score = -1.86, R) >droAna3.scaffold_12984 232316 84 - 754457 ------------------AAAUACAACAUCCCCAUCCACAUUGGCAGCACUUUUACGAGUUGCCAACUACCUUGUACAACGAAUUUUUAUUACUGCACUAGG ------------------......................((((((((.(......).))))))))...(((.((.((...(((....)))..)).)).))) ( -13.40, z-score = -1.94, R) >droMoj3.scaffold_6500 20978327 95 + 32352404 -----GACACUGAGGUGAAGGAAGAGCAAGUGUAACG--AUUGGCAACAAUUUUACGAGUCGCUAGCUACUUUUUACAACGAAUUUUCAUUAUUGCACAAGG -----....((...(((.(((((((((.((((...((--((((....))))....))...)))).))).)))))).....((....)).......))).)). ( -18.10, z-score = -0.45, R) >droGri2.scaffold_15126 5769523 100 + 8399593 GAGGUCCAAGUAAAGAGCAACAAAAGCAUUUGAAGCU--AUUGGCAACAAUUUUACGAGUUGCCAACUACUUUUUACAACGAAUUUUCAUUAUUGCAAGAGG .....((..((((((((.......(((.......)))--.((((((((..........))))))))...))))))))......((((((....)).)))))) ( -19.60, z-score = -1.42, R) >droVir3.scaffold_12963 12256834 89 + 20206255 -----------GAACAGCGACAAAAACAAUUGAAACG--AUUGGCAACACUUUUACGAGUUGCCAUCUACUUUUUACAACGAAUUUUCAUUAUAGCACAAGG -----------.....((..(((......)))....(--((.((((((.(......).)))))))))...........................))...... ( -12.90, z-score = -1.84, R) >consensus __AAACCUAC__AA_CAGCAG__CAGCAGCCACAUCAACAUUGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUUUAUUAUUGCACAAGG ...........................................(((((..........)))))......((((((.(((..(.......)..))).)))))) ( -8.90 = -9.01 + 0.11)
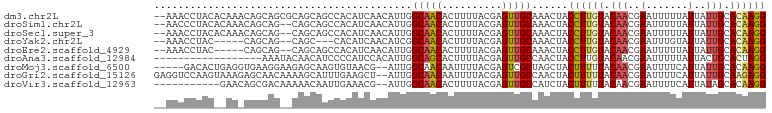
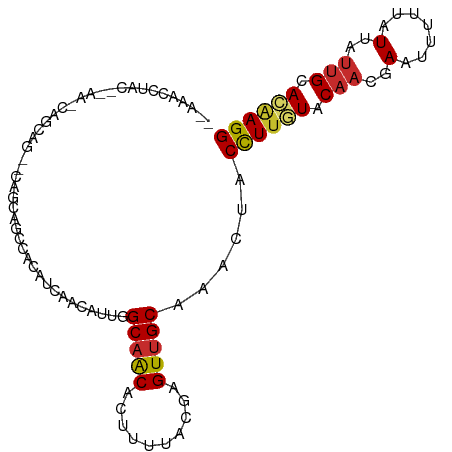
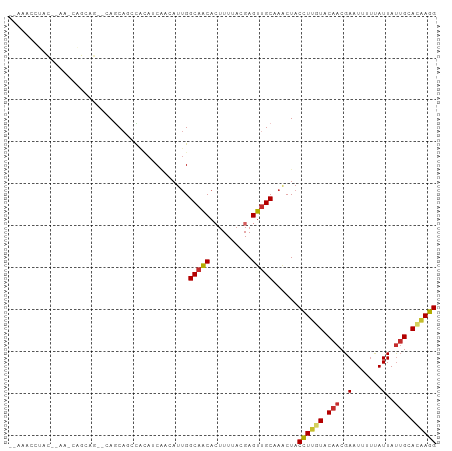
| Location | 15,554,931 – 15,555,031 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 74.71 |
| Shannon entropy | 0.53527 |
| G+C content | 0.39681 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -10.82 |
| Energy contribution | -11.49 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.891652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15554931 100 - 23011544 CCUUGUGCAAUAAUAAAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCAAUGUUGAUGUGGCUGCUGCGCUGCUGUUUGUGUAGGUUU-- ((((((((((((((....))).)))))))))))..(((.(((((.(......).))))).))).............((((((........))))))....-- ( -28.00, z-score = -1.82, R) >droSim1.chr2L 15312987 98 - 22036055 CCUUGUGCAAUAAUAAAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCAAUGUUGAUGUGGCUGCUG--CUGCUGUUUGUGUAGGGUU-- ((((((((((((((....))).)))))))))))..(((.(((((.(......).))))).)))......(..((....)--)..)...............-- ( -26.00, z-score = -1.47, R) >droSec1.super_3 1816783 98 - 7220098 CCUUGUGCAAUAAUAAAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCAAUGUUGAUGUGGCUGCUG--CUGCUGUUUGUGUAGGUUU-- ((((((((((((((....))).)))))))))))..(((.(((((.(......).))))).)))......(..((....)--)..)...............-- ( -26.00, z-score = -1.67, R) >droYak2.chr2L 2554177 90 + 22324452 CCUUGUGCAAUAAUACAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCGAUGUUGAUGUG---GCUG--CUGCUG-----GUAGGUUU-- ((((((((((((((....))).)))))))))))..(((.(((((.(......).))))).))).........---.(((--(.....-----))))....-- ( -24.30, z-score = -1.19, R) >droEre2.scaffold_4929 7303679 93 - 26641161 CCUUGUGCAAUAAUAAAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCAAUGUUGAUGUGGCUGCUG--CUGCUG-----GUAGGUUU-- ((((((((((((((....))).)))))))))))..(((.(((((.(......).))))).)))......(..((....)--)..)..-----........-- ( -26.00, z-score = -1.84, R) >droAna3.scaffold_12984 232316 84 + 754457 CCUAGUGCAGUAAUAAAAAUUCGUUGUACAAGGUAGUUGGCAACUCGUAAAAGUGCUGCCAAUGUGGAUGGGGAUGUUGUAUUU------------------ ((((((((((..(.......)..))))))......(((((((.(.(......).).))))))).....))))............------------------ ( -19.00, z-score = -0.75, R) >droMoj3.scaffold_6500 20978327 95 - 32352404 CCUUGUGCAAUAAUGAAAAUUCGUUGUAAAAAGUAGCUAGCGACUCGUAAAAUUGUUGCCAAU--CGUUACACUUGCUCUUCCUUCACCUCAGUGUC----- ....(.((((((((((....))))))).....(((((..(((((..........)))))....--.)))))...))).)......(((....)))..----- ( -15.60, z-score = -0.89, R) >droGri2.scaffold_15126 5769523 100 - 8399593 CCUCUUGCAAUAAUGAAAAUUCGUUGUAAAAAGUAGUUGGCAACUCGUAAAAUUGUUGCCAAU--AGCUUCAAAUGCUUUUGUUGCUCUUUACUUGGACCUC ((..((((((((((....))).))))))).((((((((((((((..........)))))))))--(((..((((....))))..)))...)))))))..... ( -26.00, z-score = -3.90, R) >droVir3.scaffold_12963 12256834 89 - 20206255 CCUUGUGCUAUAAUGAAAAUUCGUUGUAAAAAGUAGAUGGCAACUCGUAAAAGUGUUGCCAAU--CGUUUCAAUUGUUUUUGUCGCUGUUC----------- .(((....((((((((....))))))))..)))..(((((((((.(......).)))))).))--)((..(((......)))..)).....----------- ( -19.40, z-score = -2.75, R) >consensus CCUUGUGCAAUAAUAAAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCAAUGUUGAUGUGGCUGCUG__CUGCUG_UU__GUAGGUUU__ ..((((((((((((....))).)))))))))....(((.(((((.(......).))))).)))....................................... (-10.82 = -11.49 + 0.67)
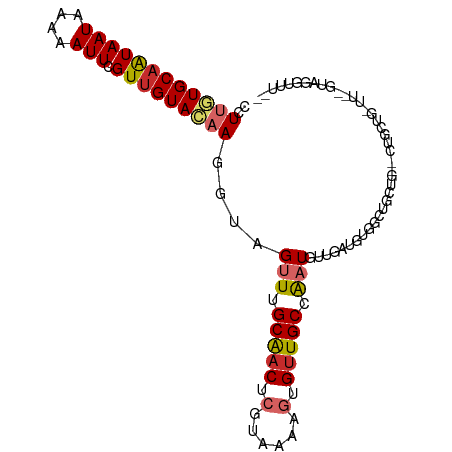
| Location | 15,554,944 – 15,555,043 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.86 |
| Shannon entropy | 0.37346 |
| G+C content | 0.41667 |
| Mean single sequence MFE | -21.99 |
| Consensus MFE | -12.77 |
| Energy contribution | -12.26 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
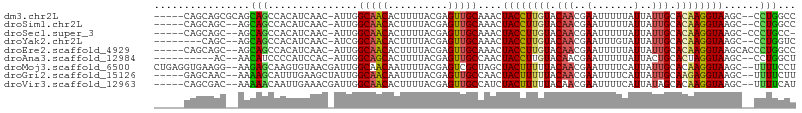
>dm3.chr2L 15554944 99 + 23011544 -----CAGCAGCGCAGCAGCCACAUCAAC-AUUGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUUUAUUAUUGCACAAGGUAAGC--CCUGGCC -----(((..(((((((.((((.......-..))))....(......).)))))....((((((((.(((..(.......)..))).)))))))).))--.)))... ( -25.10, z-score = -1.78, R) >droSim1.chr2L 15313000 97 + 22036055 -----CAGCAGC--AGCAGCCACAUCAAC-AUUGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUUUAUUAUUGCACAAGGUAAGC--CCUGGCC -----..((...--.)).((((.......-....(((((.(......).)))))....((((((((.(((..(.......)..))).))))))))...--..)))). ( -23.50, z-score = -1.71, R) >droSec1.super_3 1816796 97 + 7220098 -----CAGCAGC--AGCAGCCACAUCAAC-AUUGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUUUAUUAUUGCACAAGGUAAGC-CCCUGCC- -----..((((.--.((............-....(((((.(......).)))))....((((((((.(((..(.......)..))).)))))))).))-..)))).- ( -25.30, z-score = -2.83, R) >droYak2.chr2L 2554185 94 - 22324452 --------CAGC--AGCAGCCACAUCAAC-AUCGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUGUAUUAUUGCACAAGGUAAGC--CCUGGUC --------(((.--.((............-....(((((.(......).)))))....((((((((.(((..(.......)..))).)))))))).))--.)))... ( -21.00, z-score = -1.06, R) >droEre2.scaffold_4929 7303687 99 + 26641161 -----CAGCAGC--AGCAGCCACAUCAAC-AUUGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUUUAUUAUUGCACAAGGUAAGCACCCUGGCC -----..((.((--(((.((((.......-..))))....(......).)))))....((((((((.(((..(.......)..))).)))))))).))......... ( -23.60, z-score = -1.73, R) >droAna3.scaffold_12984 232320 92 - 754457 ----------AC--AACAUCCCCAUCCAC-AUUGGCAGCACUUUUACGAGUUGCCAACUACCUUGUACAACGAAUUUUUAUUACUGCACUAGGUAAGC--CCUGGCU ----------..--...............-.((((((((.(......).)))))))).(((((.((.((...(((....)))..)).)).)))))(((--....))) ( -19.40, z-score = -1.76, R) >droMoj3.scaffold_6500 20978331 103 + 32352404 CUGAGGUGAAGG--AAGAGCAAGUGUAACGAUUGGCAACAAUUUUACGAGUCGCUAGCUACUUUUUACAACGAAUUUUCAUUAUUGCACAAGGUAAGC--UUUUCCU .........(((--((((((...(((((.(((((....))))))))))..........((((((...(((..(.......)..)))...)))))).))--))))))) ( -23.50, z-score = -0.97, R) >droGri2.scaffold_15126 5769537 98 + 8399593 -----GAGCAAC--AAAAGCAUUUGAAGCUAUUGGCAACAAUUUUACGAGUUGCCAACUACUUUUUACAACGAAUUUUCAUUAUUGCAAGAGGUAAGC--UUUUCUU -----..((...--....))....((((((.((((((((..........)))))))).((((((((.(((..(.......)..))).)))))))))))--))).... ( -22.60, z-score = -2.58, R) >droVir3.scaffold_12963 12256837 98 + 20206255 -----CAGCGAC--AAAAACAAUUGAAACGAUUGGCAACACUUUUACGAGUUGCCAUCUACUUUUUACAACGAAUUUUCAUUAUAGCACAAGGUAAGC--UUUUCAU -----.(((...--...............(((.((((((.(......).)))))))))(((((((((.((.((....)).)).)))...)))))).))--)...... ( -13.90, z-score = -0.80, R) >consensus _____CAGCAGC__AGCAGCCACAUCAAC_AUUGGCAACACUUUUACGAGUUGCAAACUACCUUGUACAACGAAUUUUUAUUAUUGCACAAGGUAAGC__CCUGGCC ................(((...............(((((..........)))))....((((((((.(((..(.......)..))).))))))))......)))... (-12.77 = -12.26 + -0.52)
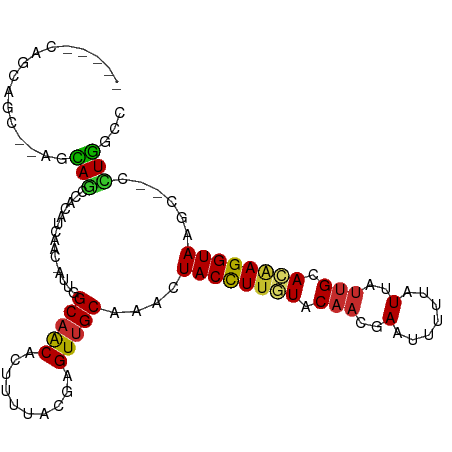
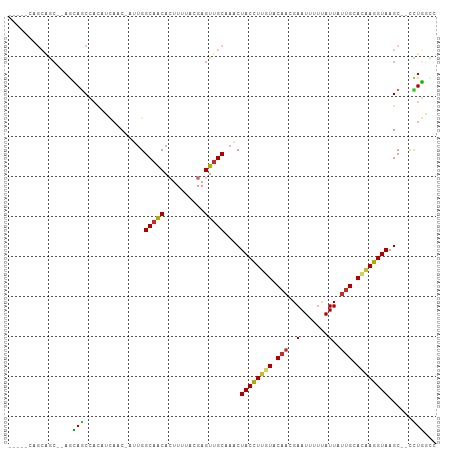
| Location | 15,554,944 – 15,555,043 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.86 |
| Shannon entropy | 0.37346 |
| G+C content | 0.41667 |
| Mean single sequence MFE | -29.29 |
| Consensus MFE | -13.91 |
| Energy contribution | -14.48 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.884729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 15554944 99 - 23011544 GGCCAGG--GCUUACCUUGUGCAAUAAUAAAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCAAU-GUUGAUGUGGCUGCUGCGCUGCUG----- (((((((--(((((((((((((((((((....))).)))))))))))))(((.(((((.(......).))))).)))-........)))).))).)))....----- ( -33.90, z-score = -2.41, R) >droSim1.chr2L 15313000 97 - 22036055 GGCCAGG--GCUUACCUUGUGCAAUAAUAAAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCAAU-GUUGAUGUGGCUGCU--GCUGCUG----- (((((((--(((.(((((((((((((((....))).)))))))))))))))))(((((.(......).)))))....-.......)))))((.--...))..----- ( -32.90, z-score = -2.62, R) >droSec1.super_3 1816796 97 - 7220098 -GGCAGGG-GCUUACCUUGUGCAAUAAUAAAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCAAU-GUUGAUGUGGCUGCU--GCUGCUG----- -(((((.(-(((((((((((((((((((....))).)))))))))))))(((.(((((.(......).))))).)))-........)))).))--)))....----- ( -37.70, z-score = -4.16, R) >droYak2.chr2L 2554185 94 + 22324452 GACCAGG--GCUUACCUUGUGCAAUAAUACAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCGAU-GUUGAUGUGGCUGCU--GCUG-------- ...((((--(((((((((((((((((((....))).)))))))))))))(((.(((((.(......).))))).)))-........)))).))--)...-------- ( -29.80, z-score = -2.13, R) >droEre2.scaffold_4929 7303687 99 - 26641161 GGCCAGGGUGCUUACCUUGUGCAAUAAUAAAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCAAU-GUUGAUGUGGCUGCU--GCUGCUG----- (((((.(..((.((((((((((((((((....))).)))))))))))))(((.(((((.(......).))))).)))-))...).)))))((.--...))..----- ( -33.70, z-score = -2.67, R) >droAna3.scaffold_12984 232320 92 + 754457 AGCCAGG--GCUUACCUAGUGCAGUAAUAAAAAUUCGUUGUACAAGGUAGUUGGCAACUCGUAAAAGUGCUGCCAAU-GUGGAUGGGGAUGUU--GU---------- ..(((..--((.(((((.((((((..(.......)..)))))).)))))(((((((.(.(......).).)))))))-))...))).......--..---------- ( -25.10, z-score = -0.72, R) >droMoj3.scaffold_6500 20978331 103 - 32352404 AGGAAAA--GCUUACCUUGUGCAAUAAUGAAAAUUCGUUGUAAAAAGUAGCUAGCGACUCGUAAAAUUGUUGCCAAUCGUUACACUUGCUCUU--CCUUCACCUCAG (((((.(--((........(((((((((....))).))))))....(((((..(((((..........))))).....)))))....))).))--)))......... ( -22.20, z-score = -2.04, R) >droGri2.scaffold_15126 5769537 98 - 8399593 AAGAAAA--GCUUACCUCUUGCAAUAAUGAAAAUUCGUUGUAAAAAGUAGUUGGCAACUCGUAAAAUUGUUGCCAAUAGCUUCAAAUGCUUUU--GUUGCUC----- ((.((((--((.......((((((((((....))).))))))).((((.(((((((((..........))))))))).)))).....))))))--.))....----- ( -27.50, z-score = -4.30, R) >droVir3.scaffold_12963 12256837 98 - 20206255 AUGAAAA--GCUUACCUUGUGCUAUAAUGAAAAUUCGUUGUAAAAAGUAGAUGGCAACUCGUAAAAGUGUUGCCAAUCGUUUCAAUUGUUUUU--GUCGCUG----- ...((((--((....(((....((((((((....))))))))..)))..(((((((((.(......).)))))).))).........))))))--.......----- ( -20.80, z-score = -1.96, R) >consensus AGCCAGG__GCUUACCUUGUGCAAUAAUAAAAAUUCGUUGUACAAGGUAGUUUGCAACUCGUAAAAGUGUUGCCAAU_GUUGAUGUGGCUGCU__GCUGCUG_____ .........((((((.((((((((((((....))).))))))))).))))...(((((.(......).)))))..............)).................. (-13.91 = -14.48 + 0.57)

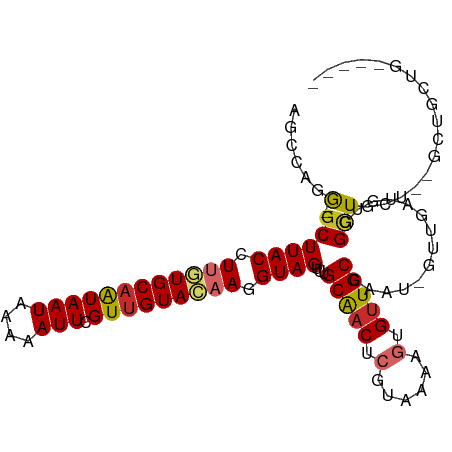
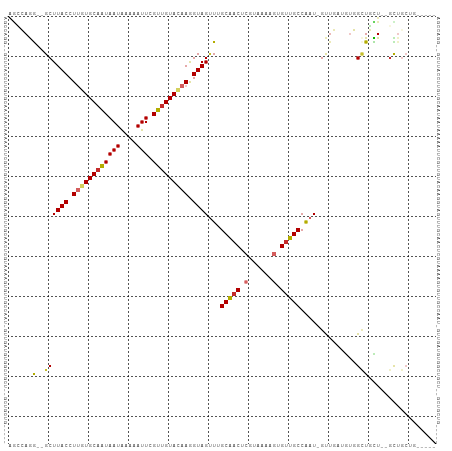
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:12 2011