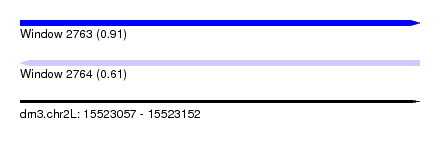
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 15,523,057 – 15,523,152 |
| Length | 95 |
| Max. P | 0.912771 |

| Location | 15,523,057 – 15,523,152 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.84 |
| Shannon entropy | 0.51942 |
| G+C content | 0.51270 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -15.97 |
| Energy contribution | -16.27 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
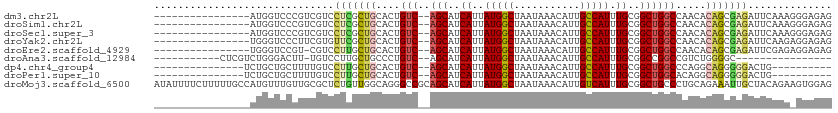
>dm3.chr2L 15523057 95 + 23011544 ----------------AUGGUCCCGUCGUCCUCGCUGCACUGUC--AGCAUCAUUAUGGCUAAUAAACAUUGCCAUUUGCGGCUGGCCAACACAGCGAGAUUCAAAGGGAGAG ----------------....((((......(((((((....(((--(((..((..(((((...........))))).))..)))))).....))))))).......))))... ( -34.02, z-score = -2.64, R) >droSim1.chr2L 15280654 95 + 22036055 ----------------AUGGUCCCGUCGUCCUCGCUGCACUGUC--AGCAUCAUUAUGGCUAAUAAACAUUGCCAUUUGCGGCUGGCCAACACAGCGAGAUUCAAAGGGAGAG ----------------....((((......(((((((....(((--(((..((..(((((...........))))).))..)))))).....))))))).......))))... ( -34.02, z-score = -2.64, R) >droSec1.super_3 1776106 95 + 7220098 ----------------AUGGUCCCGUCGUCCUCGCUGCACUGUC--AGCAUCAUUAUGGCUAAUAAACAUUGCCAUUUGCGGCUGGCCAACACAGCGAGAUUCAAAGGGAGAG ----------------....((((......(((((((....(((--(((..((..(((((...........))))).))..)))))).....))))))).......))))... ( -34.02, z-score = -2.64, R) >droYak2.chr2L 2518008 95 - 22324452 ----------------UGGGUCCCUUCGUGUUCGCUGCACUGUC--AGCAUCAUUAUGGCUAAUAAACAUUGCCAUUUGCGGCUGGCCAACACAGCGAGAUUCAAGAGGAGAG ----------------....((((((....(((((((....(((--(((..((..(((((...........))))).))..)))))).....)))))))....))).)))... ( -31.30, z-score = -1.61, R) >droEre2.scaffold_4929 7268999 94 + 26641161 ----------------UGGGUCCGU-CGUCCUUGCUGCACUGUC--AGCAUCAUUAUGGCUAAUAAACAUUGCCAUUUGCGGCUGGCCAACACAGCGAGAUUCGAGAGGAGAG ----------------....(((.(-((..(((((((....(((--(((..((..(((((...........))))).))..)))))).....)))))))...)))..)))... ( -32.40, z-score = -1.71, R) >droAna3.scaffold_12984 116155 83 - 754457 -----------CUCGUCUGGGACUU-UGUCCUUGCUGCCCUGUC--AGCAUCAUUAUGGCUAAUAAACAUUGCCAUUUGCGGCCGGCCGUCUGGGGC---------------- -----------...((..(((((..-.))))).)).(((((...--.(((.....(((((...........))))).)))((....))....)))))---------------- ( -24.20, z-score = 0.15, R) >dp4.chr4_group4 4303528 86 - 6586962 ---------------UCUGCUGCUUUUGUCCUUGCUGCACUGUC--AGCAUCAUUAUGGCUAAUAAACAUUGCCAUUUGCGGCUGGCCCAGGCAGGGGGACUG---------- ---------------............((((((.((((...(((--(((..((..(((((...........))))).))..))))))....))))))))))..---------- ( -30.50, z-score = -1.09, R) >droPer1.super_10 2139998 86 - 3432795 ---------------UCUGCUGCUUUUGUCCUUGCUGCACUGUC--AGCAUCAUUAUGGCUAAUAAACAUUGCCAUUUGCGGCUGGCACAGGCAGGGGGACUG---------- ---------------............((((((.((((..((((--(((..((..(((((...........))))).))..)))))))...))))))))))..---------- ( -32.30, z-score = -2.17, R) >droMoj3.scaffold_6500 20929385 113 + 32352404 AUAUUUUCUUUUUGCCAUGUUUGUUGCGCUCUGUUGGCAGGGCCGCAGCAUCAUUAUGGCUAAUAAACAUUGUCAUUUGCGGCUGCCCUGCAGAAAUUGCUACAGAAGUGGAG ..............((((.(((((.(((.....((.(((((((.((.(((.....(((((...........))))).))).)).))))))).))...))).))))).)))).. ( -37.20, z-score = -1.55, R) >consensus ________________AUGGUCCCUUCGUCCUCGCUGCACUGUC__AGCAUCAUUAUGGCUAAUAAACAUUGCCAUUUGCGGCUGGCCAACACAGCGAGAUUCAAA_GGAGAG ..............................(((((((....(((..(((..((..(((((...........))))).))..)))))).....))))))).............. (-15.97 = -16.27 + 0.30)

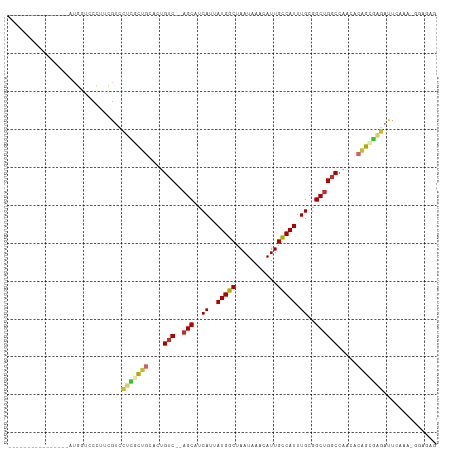
| Location | 15,523,057 – 15,523,152 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.84 |
| Shannon entropy | 0.51942 |
| G+C content | 0.51270 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -11.41 |
| Energy contribution | -11.86 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
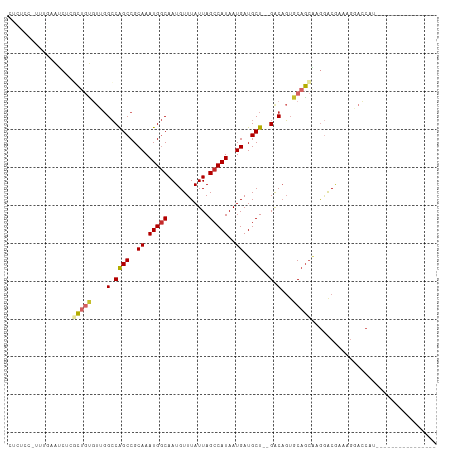
>dm3.chr2L 15523057 95 - 23011544 CUCUCCCUUUGAAUCUCGCUGUGUUGGCCAGCCGCAAAUGGCAAUGUUUAUUAGCCAUAAUGAUGCU--GACAGUGCAGCGAGGACGACGGGACCAU---------------- ...((((.(((..((((((((..(((..((((..((.(((((...........)))))..))..)))--).)))..)))))))).))).))))....---------------- ( -38.70, z-score = -3.84, R) >droSim1.chr2L 15280654 95 - 22036055 CUCUCCCUUUGAAUCUCGCUGUGUUGGCCAGCCGCAAAUGGCAAUGUUUAUUAGCCAUAAUGAUGCU--GACAGUGCAGCGAGGACGACGGGACCAU---------------- ...((((.(((..((((((((..(((..((((..((.(((((...........)))))..))..)))--).)))..)))))))).))).))))....---------------- ( -38.70, z-score = -3.84, R) >droSec1.super_3 1776106 95 - 7220098 CUCUCCCUUUGAAUCUCGCUGUGUUGGCCAGCCGCAAAUGGCAAUGUUUAUUAGCCAUAAUGAUGCU--GACAGUGCAGCGAGGACGACGGGACCAU---------------- ...((((.(((..((((((((..(((..((((..((.(((((...........)))))..))..)))--).)))..)))))))).))).))))....---------------- ( -38.70, z-score = -3.84, R) >droYak2.chr2L 2518008 95 + 22324452 CUCUCCUCUUGAAUCUCGCUGUGUUGGCCAGCCGCAAAUGGCAAUGUUUAUUAGCCAUAAUGAUGCU--GACAGUGCAGCGAACACGAAGGGACCCA---------------- ...(((..(((....((((((..(((..((((..((.(((((...........)))))..))..)))--).)))..))))))...)))..)))....---------------- ( -33.50, z-score = -2.61, R) >droEre2.scaffold_4929 7268999 94 - 26641161 CUCUCCUCUCGAAUCUCGCUGUGUUGGCCAGCCGCAAAUGGCAAUGUUUAUUAGCCAUAAUGAUGCU--GACAGUGCAGCAAGGACG-ACGGACCCA---------------- ...(((..(((..(((.((((..(((..((((..((.(((((...........)))))..))..)))--).)))..)))).))).))-).)))....---------------- ( -30.00, z-score = -1.45, R) >droAna3.scaffold_12984 116155 83 + 754457 ----------------GCCCCAGACGGCCGGCCGCAAAUGGCAAUGUUUAUUAGCCAUAAUGAUGCU--GACAGGGCAGCAAGGACA-AAGUCCCAGACGAG----------- ----------------(((((((.(((....)))...(((((...........))))).......))--)...)))).....((((.-..))))........----------- ( -21.30, z-score = 0.04, R) >dp4.chr4_group4 4303528 86 + 6586962 ----------CAGUCCCCCUGCCUGGGCCAGCCGCAAAUGGCAAUGUUUAUUAGCCAUAAUGAUGCU--GACAGUGCAGCAAGGACAAAAGCAGCAGA--------------- ----------..((((..(((((((...((((..((.(((((...........)))))..))..)))--).))).))))...))))............--------------- ( -26.80, z-score = -0.73, R) >droPer1.super_10 2139998 86 + 3432795 ----------CAGUCCCCCUGCCUGUGCCAGCCGCAAAUGGCAAUGUUUAUUAGCCAUAAUGAUGCU--GACAGUGCAGCAAGGACAAAAGCAGCAGA--------------- ----------..((((..((((((((..((((..((.(((((...........)))))..))..)))--))))).))))...))))............--------------- ( -27.30, z-score = -1.65, R) >droMoj3.scaffold_6500 20929385 113 - 32352404 CUCCACUUCUGUAGCAAUUUCUGCAGGGCAGCCGCAAAUGACAAUGUUUAUUAGCCAUAAUGAUGCUGCGGCCCUGCCAACAGAGCGCAACAAACAUGGCAAAAAGAAAAUAU ..(((....(((.((...(((((((((((.((.(((...........((((((....))))))))).)).)))))))....)))).)).)))....))).............. ( -27.20, z-score = 0.07, R) >consensus CUCUCC_UUUGAAUCUCGCUGUGUUGGCCAGCCGCAAAUGGCAAUGUUUAUUAGCCAUAAUGAUGCU__GACAGUGCAGCAAGGACGAAAGGACCAU________________ .................(((((....((((........))))..((((....(((.........)))..))))..)))))................................. (-11.41 = -11.86 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:43:02 2011